Group-level Enrichment Analysis (brainenrich)
brainenrich_demo.RmdIntroduction
This tutorial demonstrates how to perform an enrichment analysis of derived group-level statistics using the BrainEnrich package. We will use effect size maps of Bipolar Disorders (BD) and gene expression data to identify significant gene sets.
1. Prepare Data for the Analysis
1.1. Load brain_data (effect size maps of Bipolar Disorders)
We will load brain data that contains the effect size maps of Bipolar Disorders. This data includes case-control comparisons of regional cortical thickness.
# Load brain data from the package
data(brain_data)
# Display the structure of the brain data
str(brain_data)
#> 'data.frame': 34 obs. of 1 variable:
#> $ MDD: num -0.058 -0.042 -0.014 0.047 -0.041 -0.117 -0.063 -0.049 -0.104 -0.023 ...Show effect size map for BD
Visualize the effect size map to understand the regional differences in cortical thickness.
First, we define a plot function (this is not included in the package as it is not a core function).
#' Plot Brain Data
#'
#' This function creates a brain plot using the ggplot2 and ggseg packages.
#'
#' df2plot A data frame containing the data to plot.
#' ats A character string indicating the atlas to use ('dx', 'dk', 'aseg').
#' what2plot A character string indicating the variable to plot ('statistic').
#' filterby A character string indicating the filter to apply ('p.value', 'p.adj', 'none').
#' title2show A character string indicating the title of the plot.
#' limit2show A numeric vector of length 2 indicating the limits for the color scale.
#' legend2show A character string indicating the legend title.
#' hide_legend A logical value indicating whether to hide the legend.
#' hem A character string indicating which hemisphere to plot ('both', 'left', 'right').
#' low A character string indicating the color for the low end of the scale.
#' mid A character string indicating the color for the midpoint of the scale.
#' high A character string indicating the color for the high end of the scale.
#' sufix2remove A character string indicating the suffix to remove from labels.
#' return A ggplot2 object.
plot_brain <- function(df2plot,
ats = c("dx", "dk", "aseg"),
what2plot = "statistic",
filterby = c("p.value", "p.adj", "none"),
title2show = "",
limit2show = c(-15, 15),
legend2show = "Stat",
hide_legend = FALSE,
hem = "both",
low = "steelblue1",
mid = "white",
high = "firebrick1",
sufix2remove = "_thickavg") {
# Match arguments with allowed values
ats <- match.arg(ats)
filterby <- match.arg(filterby)
# Apply filters based on p-values
if (filterby == "p.value") {
df2plot <- df2plot %>% filter(.data$p.value < 0.05)
} else if (filterby == "p.adj") {
df2plot <- df2plot %>% filter(.data$p.adj < 0.05)
} else if (filterby == "none") {
df2plot <- df2plot
}
# Modify labels and join data based on anatomical terms
if (ats == "dx" || ats == "dk") {
atlas <- ifelse(ats == "dx", "desterieux", "dk")
atlas_data <- getExportedValue("ggseg", atlas)
df2plot <- df2plot %>%
mutate(
label = sub(".*L_", "lh_", .data$label),
label = sub(".*R_", "rh_", .data$label),
label = sub(sufix2remove, "", .data$label)
) %>%
brain_join(atlas_data) %>%
reposition_brain(as.formula(". ~ hemi + side"))
} else if (ats == "aseg") {
atlas_data <- getExportedValue("ggseg", "aseg") # ggseg::aseg
df2plot <- df2plot %>%
mutate(
label = sub("SV_L_", "Left-", .data$label), # Replace "SV_L_" with "Left-"
label = sub("SV_R_", "Right-", .data$label), # Replace "SV_R_" with "Right-"
label = sub("thal", "Thalamus-Proper", .data$label), # Replace "thal" with "Thalamus-Proper"
label = sub("caud", "Caudate", .data$label), # Replace "caud" with "Caudate"
label = sub("put", "Putamen", .data$label), # Replace "put" with "Putamen"
label = sub("pal", "Pallidum", .data$label), # Replace "pal" with "Pallidum"
label = sub("hippo", "Hippocampus", .data$label), # Replace "hippo" with "Hippocampus"
label = sub("amyg", "Amygdala", .data$label), # Replace "amyg" with "Amygdala"
label = sub("accumb", "Accumben-area", .data$label), # Replace "accumb" with "Accumben-area"
label = sub("LatVent", "Lateral-Ventricle", .data$label) # Replace "LatVent" with "Lateral-Ventricle"
) %>%
filter(!grepl("Accumben-area", .data$label)) %>%
brain_join(atlas_data) %>%
filter(.data$side == "coronal")
}
# Apply conditional filtering based on hemisphere
if (hem %in% c("left", "right")) {
df2plot <- df2plot %>% filter(.data$hemi == hem)
}
# Construct the plot
p <- df2plot %>%
ggplot() +
geom_sf(aes(fill = .data[[what2plot]])) +
scale_fill_gradient2(midpoint = 0, low = low, mid = mid, high = high, space = "Lab", limits = limit2show) +
ggtitle(title2show) +
theme_void() +
theme(plot.title = element_text(hjust = 0.5)) +
labs(fill = legend2show) +
{
if (hide_legend) theme(legend.position = "none")
}
return(p)
}
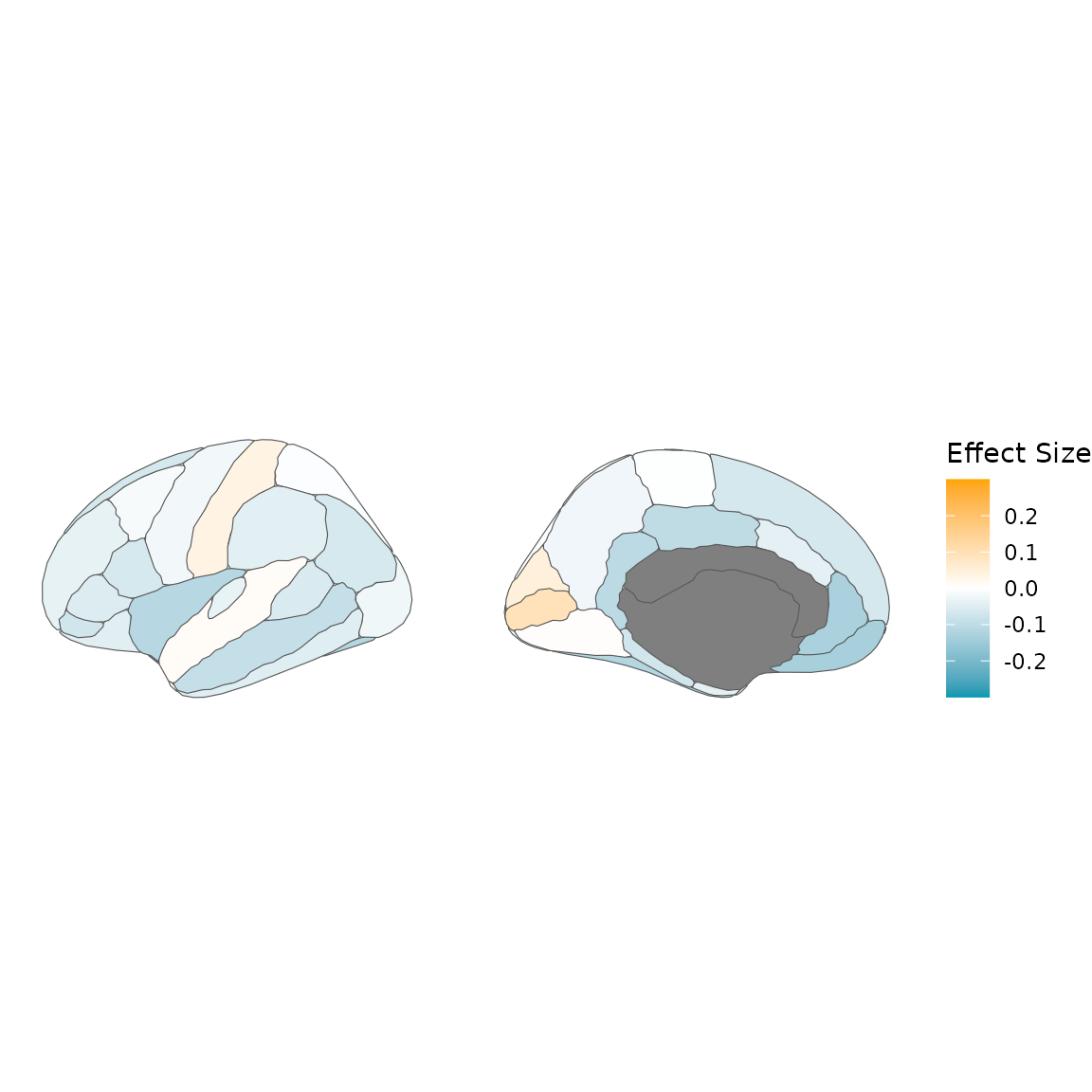
df2plot <- brain_data %>% tibble::rownames_to_column("label")
plot_brain(df2plot,
ats = "dk", filterby = "none", limit2show = c(-0.3, 0.3),
what2plot = "MDD", hem = "left", low = "#0197b2", mid = "white", high = "orange",
legend2show = "Effect Size"
)
1.2. Create perm_id for the Analysis
perm_id is a list of permutations of brain regions with a spinning approach while trying to maintain the spatial structure of the brain. It is used to generate null distributions for the enrichment analysis. If you use this function to create perm_ids, consider citing the original source at https://github.com/frantisekvasa/rotate_parcellation and relevant papers.
# read the centroid coordinates of the brain regions
data(coord_dk_lh)
perm_id_dk_lh_5000 <- rotate_parcellation(coord.l = coord_dk_lh, nrot = 5000, seed = 2024)1.3. Load gene expression data for the Analysis
gene_data <- get_geneExp(atlas = "desikan", rdonor = "r0.4", hem = "L")
str(gene_data)
#> num [1:34, 1:6513] 0.469 0.607 0.512 0.341 0.664 ...
#> - attr(*, "dimnames")=List of 2
#> ..$ : chr [1:34] "L_bankssts" "L_caudalanteriorcingulate" "L_caudalmiddlefrontal" "L_cuneus" ...
#> ..$ : chr [1:6513] "A1BG" "A1BG-AS1" "AACS" "AADAT" ...1.4. Load GO MF for the Analysis
annoData <- get_annoData(type = "GO_MF")
geneSetList <- get_geneSetList(annoData)
length(geneSetList)
#> [1] 5060
print(sprintf("Number of MF terms: %d", length(geneSetList)))
#> [1] "Number of MF terms: 5060"This step is embedded in the brainenrich function. Here is just to show how it is filtered.
selected.gs <- filter_geneSetList(bg_genes = colnames(gene_data), geneSetList = geneSetList, minGSSize = 20, maxGSSize = 200)
print(sprintf("%d MF terms have gene size ranging between 20 and 200", length(selected.gs)))2. Run the Analysis
res <- brainenrich(
brain_data = brain_data,
gene_data = gene_data,
annoData = annoData,
perm_id = perm_id_dk_lh_5000,
cor_method = "pearson",
aggre_method = "mean",
null_model = "spin_brain",
n_perm = 5000,
n_cores = 0,
minGSSize = 20,
maxGSSize = 200,
pvalueCutoff = 1,
threshold_type = "sd",
threshold = 1
)3. Visualize the Results
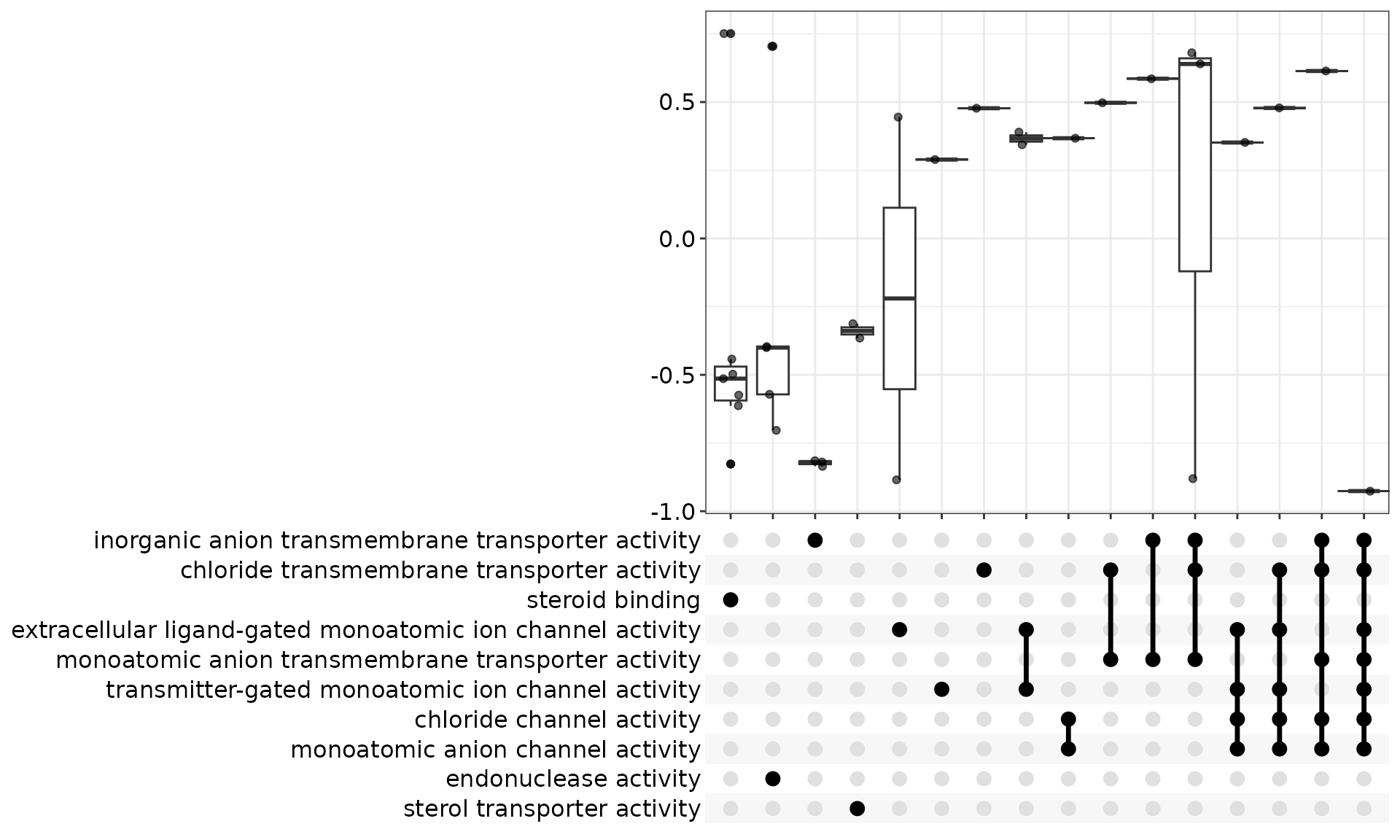
The res is a gseaResult object that is commonly used in clusterProfiler and DOSE packages. This means that we can use some existing functions (e.g., those from enrichplot package) to visualize the results.
3.2. Gene-Concept Network
geneList <- res@geneList
cnetplot(res, layout='kk', color_category='#a6a6a6', foldChange=geneList,
# color.params = list(foldChange = geneList),
showtext=TRUE,
node_label = 'gene',
showCategory =10, cex.params = list(category_node=1, gene_node=0.8, gene_label=0.8),
shadowtext = 'category')+
scale_color_gradient2(midpoint=0,low='#0197b2',mid='white',high='orange',
space ="Lab" ,limits = c(-1,1), name='Association')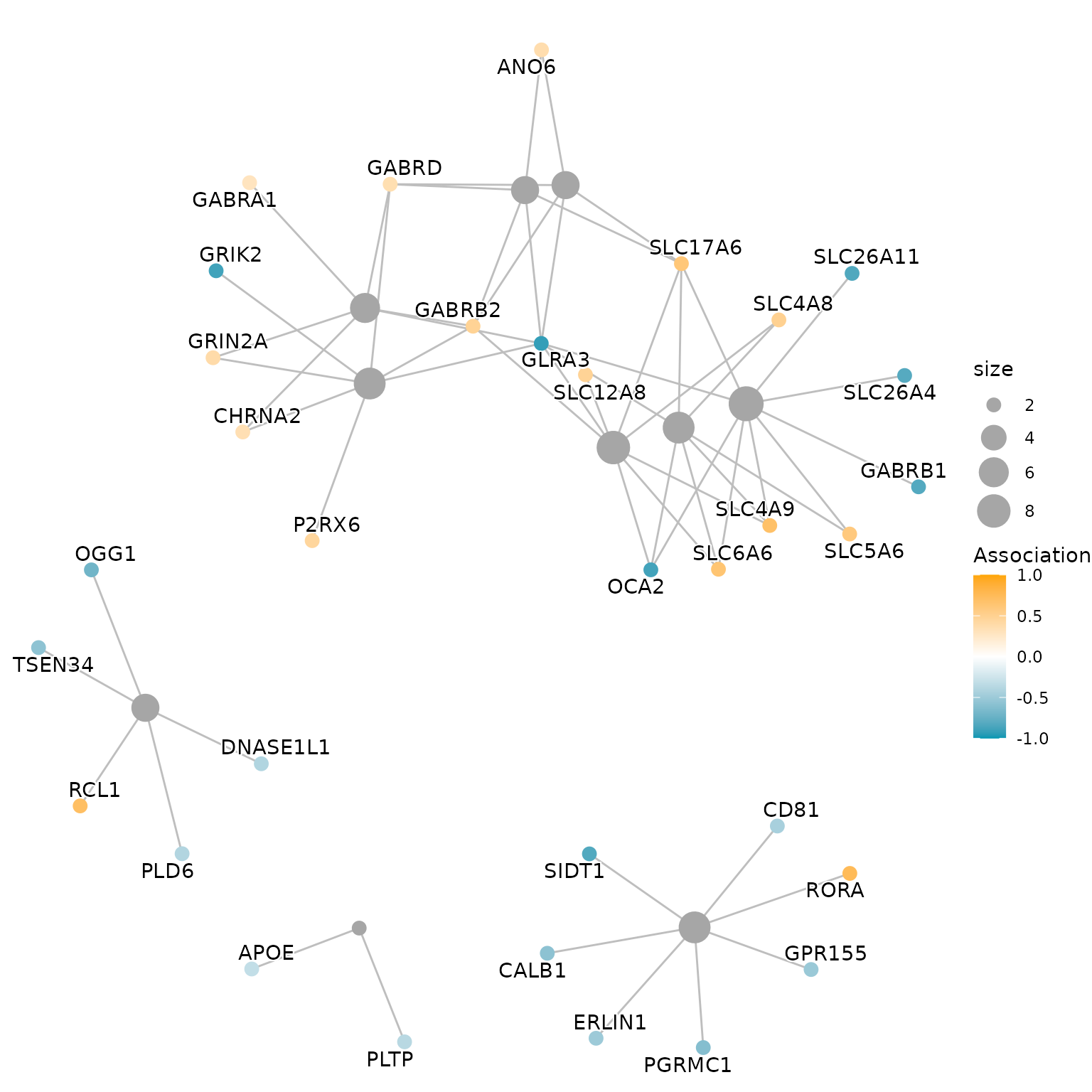
3.3. Dot Plot
dotplot(res, x = "gsScore", label_format = 50, showCategory = 30) +
xlab("GS score") +
scale_fill_gradient(high='#ffffff',low='orange',
space ="Lab" , name='p.adjusted',limits = c(0,0.05)) +
theme(legend.position = 'none')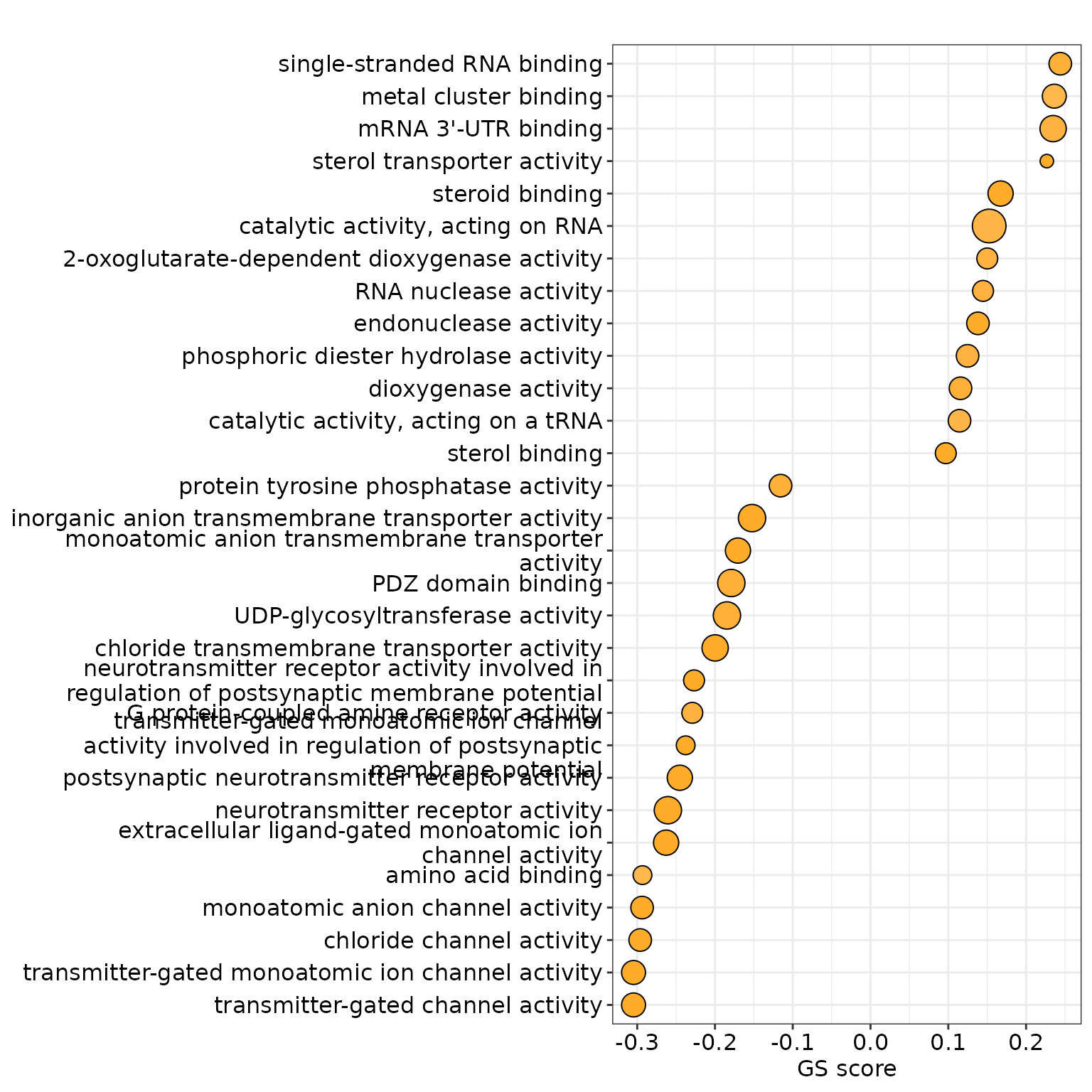
3.4. Heatplot
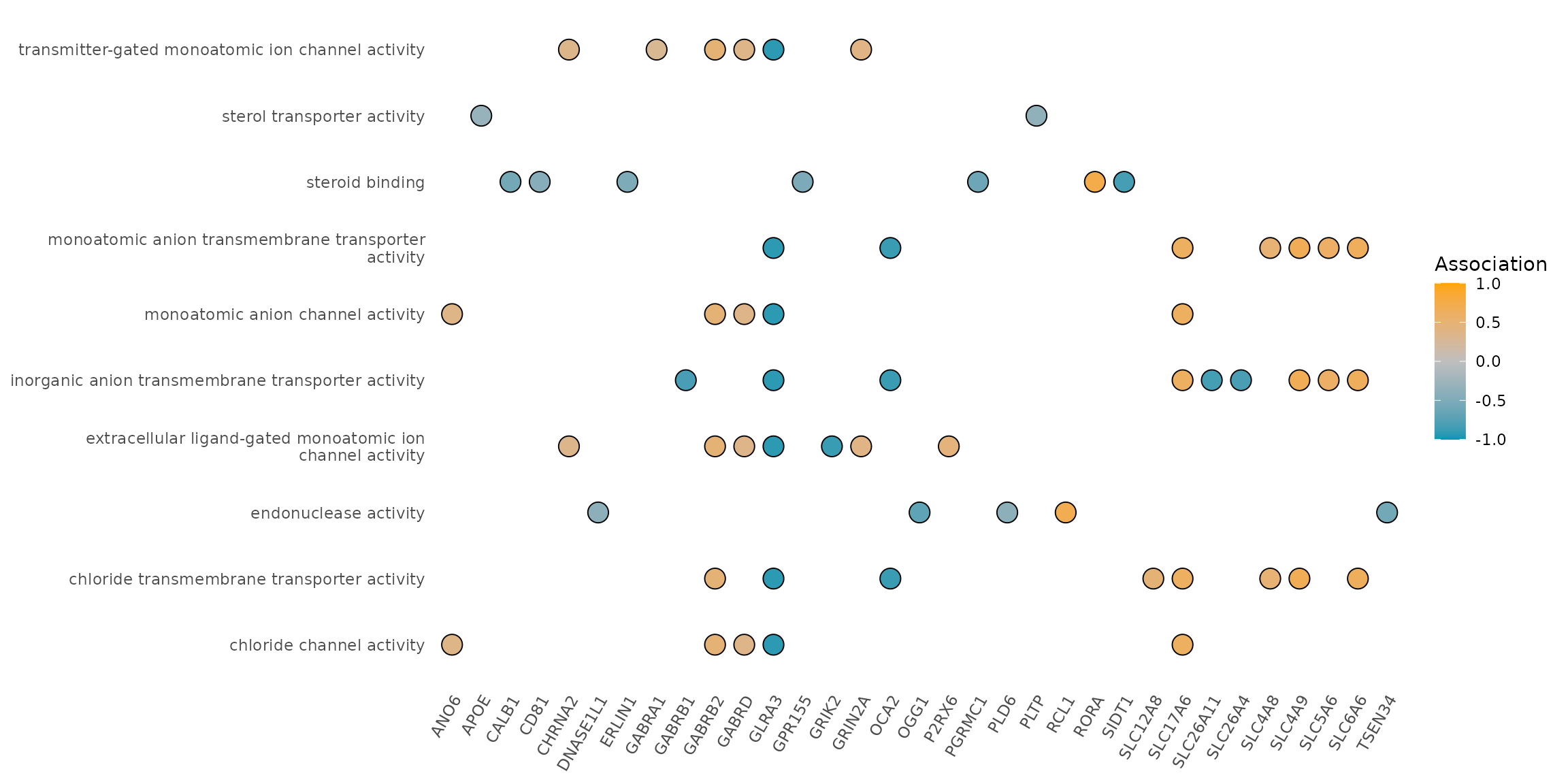
heatplot(res, foldChange = geneList, label_format = 50, showCategory = 10, symbol="dot") +
scale_fill_gradient2(
midpoint = 0, low = "#0197b2", mid = "grey", high = "orange",
space = "Lab", limits = c(-1, 1), name = "Association"
)
3.5. Tree plot based on term similarity
Note: This requires GOSemSim and org.Hs.eg.db packages.
d <- GOSemSim::godata(annoDb = "org.Hs.eg.db", ont = "MF", keytype = "SYMBOL", computeIC = FALSE) # computeIC=FALSE when use Wang method
res <- pairwise_termsim(res, method = "Wang", semData = d)
treeplot(res,
showCategory = 30,
fontsize=4,
cex_category = 0.9, color = "p.adjust",
cluster.params = list( n = 5,label_words_n = 3,
color = c("#FFB14ECC", "#89be95cc","#8dc4d2", "#99bed3cc", "#ef9d8bcc")),
offset = rel(2)
) + scale_color_continuous(high = "white", low = "orange",
space = "Lab", name = "gsScore"
)+
theme(legend.position = 'right') 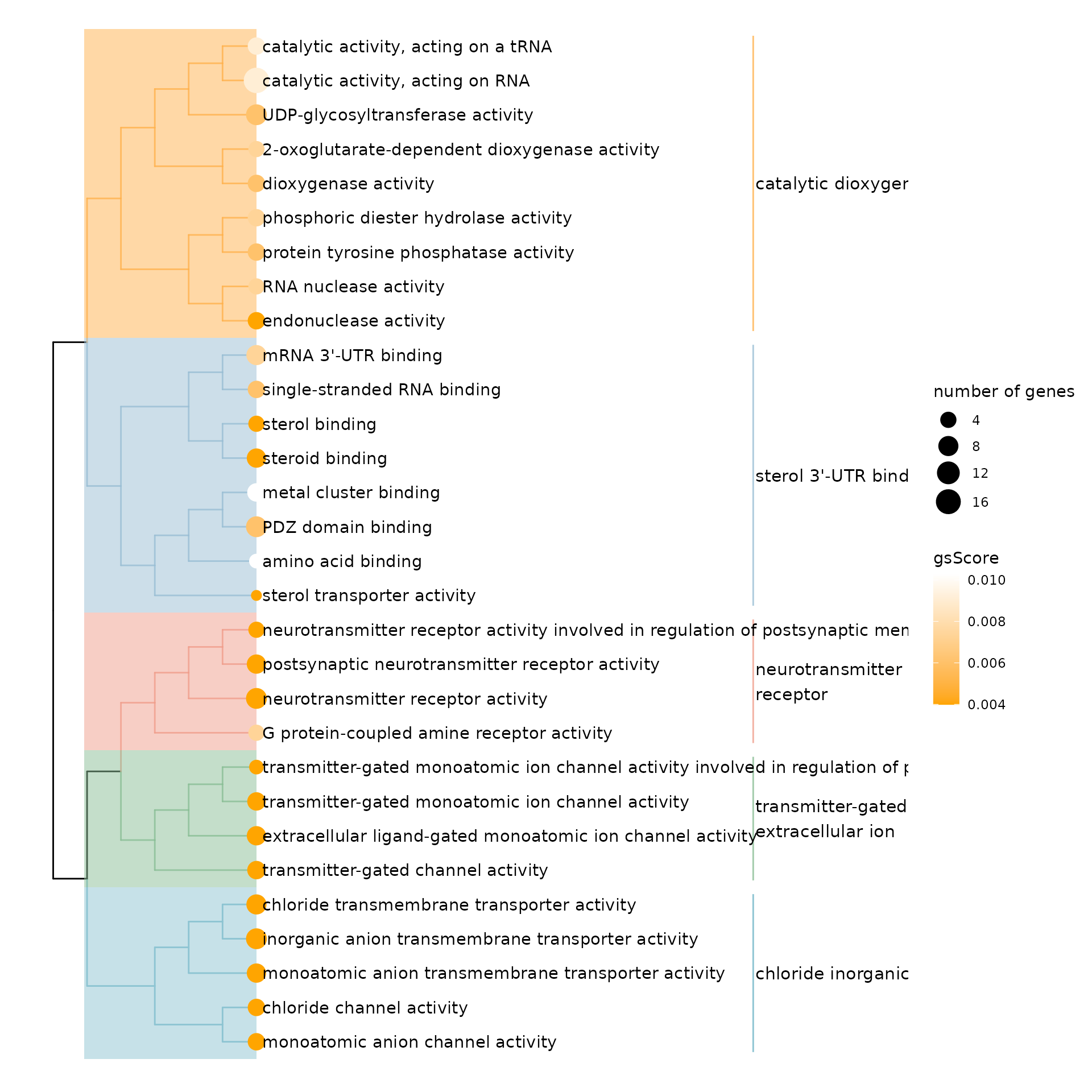
4. Put results in a Table
res.df=as.data.frame(res)
# make scollable table
kable(res.df, format = "html", escape = FALSE) %>%
kable_styling("striped", full_width = FALSE) %>%
scroll_box(width = "100%", height = "400px")| ID | Description | setSize | gsScore | pvalue | p.adjust | qvalue | core_enrichment | |
|---|---|---|---|---|---|---|---|---|
| GO:0004519 | GO:0004519 | endonuclease activity | 36 | 0.1381412 | 0.0002000 | 0.0039867 | 0.0024206 | DNASE1L1/OGG1/RCL1/TSEN34/PLD6 |
| GO:0005230 | GO:0005230 | extracellular ligand-gated monoatomic ion channel activity | 31 | -0.2628351 | 0.0002000 | 0.0039867 | 0.0024206 | CHRNA2/GABRB2/GABRD/GRIK2/GRIN2A/GLRA3/P2RX6 |
| GO:0005253 | GO:0005253 | monoatomic anion channel activity | 34 | -0.2937461 | 0.0002000 | 0.0039867 | 0.0024206 | GABRB2/GABRD/GLRA3/SLC17A6/ANO6 |
| GO:0005254 | GO:0005254 | chloride channel activity | 33 | -0.2961512 | 0.0002000 | 0.0039867 | 0.0024206 | GABRB2/GABRD/GLRA3/SLC17A6/ANO6 |
| GO:0005496 | GO:0005496 | steroid binding | 46 | 0.1672464 | 0.0002000 | 0.0039867 | 0.0024206 | CALB1/CD81/RORA/ERLIN1/PGRMC1/SIDT1/GPR155 |
| GO:0008509 | GO:0008509 | monoatomic anion transmembrane transporter activity | 47 | -0.1704597 | 0.0002000 | 0.0039867 | 0.0024206 | OCA2/SLC6A6/GLRA3/SLC5A6/SLC4A8/SLC17A6/SLC4A9 |
| GO:0015103 | GO:0015103 | inorganic anion transmembrane transporter activity | 50 | -0.1523500 | 0.0002000 | 0.0039867 | 0.0024206 | GABRB1/OCA2/SLC26A4/SLC6A6/GLRA3/SLC5A6/SLC17A6/SLC4A9/SLC26A11 |
| GO:0015108 | GO:0015108 | chloride transmembrane transporter activity | 42 | -0.1998043 | 0.0002000 | 0.0039867 | 0.0024206 | GABRB2/OCA2/SLC6A6/GLRA3/SLC4A8/SLC17A6/SLC4A9/SLC12A8 |
| GO:0015248 | GO:0015248 | sterol transporter activity | 20 | 0.2266985 | 0.0002000 | 0.0039867 | 0.0024206 | APOE/PLTP |
| GO:0022824 | GO:0022824 | transmitter-gated monoatomic ion channel activity | 29 | -0.3047262 | 0.0002000 | 0.0039867 | 0.0024206 | CHRNA2/GABRA1/GABRB2/GABRD/GRIN2A/GLRA3 |
| GO:0022835 | GO:0022835 | transmitter-gated channel activity | 29 | -0.3047262 | 0.0002000 | 0.0039867 | 0.0024206 | CHRNA2/GABRA1/GABRB2/GABRD/GRIN2A/GLRA3 |
| GO:0030594 | GO:0030594 | neurotransmitter receptor activity | 52 | -0.2605532 | 0.0002000 | 0.0039867 | 0.0024206 | ADRB1/CHRNA2/GABRB2/GABRD/GRIK2/GRIN2A/HRH2/HTR1F/GLRA3 |
| GO:0032934 | GO:0032934 | sterol binding | 32 | 0.0967745 | 0.0002000 | 0.0039867 | 0.0024206 | RORA/ERLIN1/SIDT1/GPR155 |
| GO:0098960 | GO:0098960 | postsynaptic neurotransmitter receptor activity | 32 | -0.2452023 | 0.0002000 | 0.0039867 | 0.0024206 | ADRB1/CHRNA2/GABRB1/GABRB2/GABRD/GRIK2/GRIN2A |
| GO:0099529 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential | 23 | -0.2269314 | 0.0002000 | 0.0039867 | 0.0024206 | ADRB1/GABRB2/GRIK2/GRIN2A |
| GO:1904315 | GO:1904315 | transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential | 21 | -0.2376968 | 0.0002000 | 0.0039867 | 0.0024206 | GABRB2/GRIK2/GRIN2A |
| GO:0003727 | GO:0003727 | single-stranded RNA binding | 34 | 0.2439800 | 0.0003999 | 0.0060750 | 0.0036885 | HNRNPC/POLR2G/CBX6/DHX58/PABPC1L |
| GO:0004725 | GO:0004725 | protein tyrosine phosphatase activity | 34 | -0.1157019 | 0.0003999 | 0.0060750 | 0.0036885 | DUSP3/EYA4/PTPRA/PTPRF/PGP |
| GO:0008194 | GO:0008194 | UDP-glycosyltransferase activity | 53 | -0.1846132 | 0.0003999 | 0.0060750 | 0.0036885 | EXTL2/GCNT1/MGAT1/LARGE1/XYLT1/XYLT2/B3GNT9/B3GLCT/B3GALNT2 |
| GO:0030165 | GO:0030165 | PDZ domain binding | 49 | -0.1789868 | 0.0003999 | 0.0060750 | 0.0036885 | ADRB1/ATP2B2/ATP2B3/ATP2B4/DTNA/GRIK2/SLC22A5/SSTR2/SHISA9 |
| GO:0051213 | GO:0051213 | dioxygenase activity | 33 | 0.1156888 | 0.0003999 | 0.0060750 | 0.0036885 | CDO1/P4HA2/HSPBAP1/RIOX2/TYW5 |
| GO:0003730 | GO:0003730 | mRNA 3’-UTR binding | 37 | 0.2347905 | 0.0005999 | 0.0073601 | 0.0044687 | ZFP36L1/CRYZ/HNRNPC/RPL41/ZNF385A/CSDC2/PABPC1L/CPEB2 |
| GO:0004540 | GO:0004540 | RNA nuclease activity | 34 | 0.1446817 | 0.0005999 | 0.0073601 | 0.0044687 | RCL1/CNOT7/TSEN34/DCP2 |
| GO:0008081 | GO:0008081 | phosphoric diester hydrolase activity | 36 | 0.1247722 | 0.0005999 | 0.0073601 | 0.0044687 | PDE1A/PDE2A/PLCB2/PLCB4/PLCD4 |
| GO:0008227 | GO:0008227 | G protein-coupled amine receptor activity | 23 | -0.2291764 | 0.0005999 | 0.0073601 | 0.0044687 | ADRA2A/ADRB1/HRH2/HTR1F |
| GO:0016706 | GO:0016706 | 2-oxoglutarate-dependent dioxygenase activity | 21 | 0.1501212 | 0.0005999 | 0.0073601 | 0.0044687 | P4HA2/HSPBAP1/RIOX2/TYW5 |
| GO:0140098 | GO:0140098 | catalytic activity, acting on RNA | 108 | 0.1525681 | 0.0007998 | 0.0091125 | 0.0055327 | DDX10/GATB/POLR2L/PRIM1/EMG1/METTL5/CNOT7/THG1L/DDX54/TSEN34/DHX58/TRMT2B/TDRD9/TYW5/METTL6/PTRH1/DCP2 |
| GO:0140101 | GO:0140101 | catalytic activity, acting on a tRNA | 33 | 0.1144280 | 0.0007998 | 0.0091125 | 0.0055327 | THG1L/TSEN34/TRMT2B/TYW5/PTRH1 |
| GO:0016597 | GO:0016597 | amino acid binding | 20 | -0.2932416 | 0.0009998 | 0.0102883 | 0.0062466 | GLRB/GLRA3/UBR2 |
| GO:0051540 | GO:0051540 | metal cluster binding | 20 | 0.2362941 | 0.0009998 | 0.0102883 | 0.0062466 | FXN/MOCS1/UQCRFS1/LIAS/NUBPL/AIFM3 |
| GO:0140640 | GO:0140640 | catalytic activity, acting on a nucleic acid | 155 | 0.1284692 | 0.0009998 | 0.0102883 | 0.0062466 | CHD3/DDX10/OGG1/GATB/POLR2L/PRIM1/RFC3/SMARCA1/RCL1/EMG1/METTL5/CNOT7/TDP2/THG1L/POLE4/DDX54/TSEN34/DSCC1/DHX58/TRMT2B/TDRD9/TYW5/METTL6/PTRH1/DCP2 |
| GO:0004930 | GO:0004930 | G protein-coupled receptor activity | 141 | -0.1267715 | 0.0017996 | 0.0179402 | 0.0108926 | AR/CMKLR1/CNR1/EDNRA/GABRB1/GALR1/GRM8/HRH1/HTR1F/MC1R/NTSR1/OPRM1/SSTR1/SSTR2/VIPR2/FZD6/SPHK1/GPR161/OR7E24/GPR61/QRFPR/MRGPRF/TAS2R30/OR2AK2 |
| GO:0005246 | GO:0005246 | calcium channel regulator activity | 25 | -0.1587008 | 0.0019996 | 0.0190005 | 0.0115363 | CABP1/HPCAL4/STIM2/CACNG8 |
| GO:0002039 | GO:0002039 | p53 binding | 21 | 0.0856668 | 0.0021996 | 0.0190005 | 0.0115363 | ZNF385A/SETD7/TAF3 |
| GO:0031491 | GO:0031491 | nucleosome binding | 20 | 0.1891281 | 0.0021996 | 0.0190005 | 0.0115363 | PARP1/HNRNPC/SMARCA1 |
| GO:0051219 | GO:0051219 | phosphoprotein binding | 33 | -0.1621371 | 0.0021996 | 0.0190005 | 0.0115363 | EPB41/IRS1/SNCA/MID2/SHD |
| GO:0001653 | GO:0001653 | peptide receptor activity | 34 | -0.1738875 | 0.0023995 | 0.0190005 | 0.0115363 | EDNRA/GALR1/MC1R/SSTR2/VIPR2/QRFPR |
| GO:0016741 | GO:0016741 | transferase activity, transferring one-carbon groups | 70 | 0.1644546 | 0.0023995 | 0.0190005 | 0.0115363 | HNMT/PRMT2/PNMT/EMG1/BHMT2/METTL5/METTL9/PRMT6/METTL17/TRMT2B/METTL6 |
| GO:0045309 | GO:0045309 | protein phosphorylated amino acid binding | 25 | -0.1772403 | 0.0023995 | 0.0190005 | 0.0115363 | IRS1/YWHAZ/SHD |
| GO:0004497 | GO:0004497 | monooxygenase activity | 24 | -0.2588758 | 0.0025995 | 0.0190005 | 0.0115363 | MICAL2/CYP46A1/CYP20A1/FOXRED2/CYP4Z1 |
| GO:0008186 | GO:0008186 | ATP-dependent activity, acting on RNA | 24 | 0.2141053 | 0.0025995 | 0.0190005 | 0.0115363 | DDX10/DDX54/DHX58/TDRD9 |
| GO:0015485 | GO:0015485 | cholesterol binding | 25 | 0.0960308 | 0.0025995 | 0.0190005 | 0.0115363 | ERLIN1/SIDT1/GPR155 |
| GO:0003729 | GO:0003729 | mRNA binding | 101 | 0.1162701 | 0.0027994 | 0.0190005 | 0.0115363 | GRSF1/HNRNPC/CAPRIN1/NOVA2/RPL41/SLBP/NELFE/RBM8A/CELF3/ZNF385A/CSDC2/YTHDF2/PCBP3/CELF4/CELF5/LARP4/CPEB2/HNRNPA1L2/NANOS1 |
| GO:0004879 | GO:0004879 | nuclear receptor activity | 26 | 0.1401725 | 0.0027994 | 0.0190005 | 0.0115363 | NKX3-1/THRA/THRB/NR4A3/AHRR |
| GO:0008528 | GO:0008528 | G protein-coupled peptide receptor activity | 33 | -0.1887294 | 0.0027994 | 0.0190005 | 0.0115363 | EDNRA/GALR1/MC1R/SSTR2/VIPR2/QRFPR |
| GO:0098531 | GO:0098531 | ligand-activated transcription factor activity | 26 | 0.1401725 | 0.0027994 | 0.0190005 | 0.0115363 | NKX3-1/THRA/THRB/NR4A3/AHRR |
| GO:0140993 | GO:0140993 | histone modifying activity | 71 | 0.2114749 | 0.0027994 | 0.0190005 | 0.0115363 | EYA1/EYA2/PRMT2/PRKAA2/TAF10/HDAC9/KAT7/PRMT6/RIOX2 |
| GO:0004518 | GO:0004518 | nuclease activity | 58 | 0.1236703 | 0.0029994 | 0.0195267 | 0.0118558 | OGG1/TATDN2/RCL1/CNOT7/TDP2/EXD3/TSEN34/DCP2 |
| GO:0008168 | GO:0008168 | methyltransferase activity | 68 | 0.1655802 | 0.0029994 | 0.0195267 | 0.0118558 | HNMT/PRMT2/PNMT/EMG1/BHMT2/METTL5/METTL9/PRMT6/METTL17/TRMT2B/METTL6 |
| GO:0016758 | GO:0016758 | hexosyltransferase activity | 67 | -0.1172007 | 0.0031994 | 0.0204119 | 0.0123933 | EXTL2/GCNT1/MGAT1/MTAP/B3GALNT1/LARGE1/ALG3/B4GAT1/B3GNT9/B3GALT6/B3GALNT2 |
| GO:0017022 | GO:0017022 | myosin binding | 34 | -0.1617436 | 0.0033993 | 0.0212624 | 0.0129097 | RAB11B/VEZT/TRAK2/MYO19/NKD2/CALML4 |
| GO:0003925 | GO:0003925 | G protein activity | 20 | -0.2767398 | 0.0037992 | 0.0224437 | 0.0136269 | RALB/RAB11B/RERG/RASL11A |
| GO:0008173 | GO:0008173 | RNA methyltransferase activity | 20 | 0.1472525 | 0.0037992 | 0.0224437 | 0.0136269 | EMG1/METTL5/TRMT2B/METTL6 |
| GO:0042054 | GO:0042054 | histone methyltransferase activity | 23 | 0.2547403 | 0.0037992 | 0.0224437 | 0.0136269 | PRMT2/PRMT6 |
| GO:0043178 | GO:0043178 | alcohol binding | 42 | 0.1161983 | 0.0039992 | 0.0229235 | 0.0139183 | CRABP1/RBP4/RLBP1/TRPC3/ERLIN1/SIDT1/GPR155 |
| GO:0005546 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding | 35 | -0.1017822 | 0.0041992 | 0.0229235 | 0.0139183 | SYT1/OSBPL2/RAB35/PLCB1/PHLDA3/SNX21/SYT10 |
| GO:0016278 | GO:0016278 | lysine N-methyltransferase activity | 20 | 0.3239945 | 0.0041992 | 0.0229235 | 0.0139183 | NSD2/METTL13/PRDM8/CAMKMT |
| GO:0016279 | GO:0016279 | protein-lysine N-methyltransferase activity | 20 | 0.3239945 | 0.0041992 | 0.0229235 | 0.0139183 | NSD2/METTL13/PRDM8/CAMKMT |
| GO:0016757 | GO:0016757 | glycosyltransferase activity | 105 | -0.0838306 | 0.0043991 | 0.0229235 | 0.0139183 | EXTL2/GCNT1/MTAP/UPP1/TNKS/B3GALNT1/ST3GAL5/LARGE1/ALG3/ST3GAL6/B4GAT1/SIRT4/XYLT1/XYLT2/GALNT14/ST6GALNAC5/GLT8D2/B3GNT9/B3GALT6/B3GALNT2/DPY19L2P1 |
| GO:0019212 | GO:0019212 | phosphatase inhibitor activity | 21 | -0.1521855 | 0.0043991 | 0.0229235 | 0.0139183 | MGAT5/PPP1R10/TESC/ELFN2 |
| GO:0140097 | GO:0140097 | catalytic activity, acting on DNA | 58 | 0.1277621 | 0.0043991 | 0.0229235 | 0.0139183 | CHD3/DNASE1L1/OGG1/RFC3/SMARCA1/TDP2/CHTF8/POLE4/DSCC1/PIF1 |
| GO:0003724 | GO:0003724 | RNA helicase activity | 23 | 0.1943884 | 0.0045991 | 0.0229235 | 0.0139183 | DDX10/DDX54/DHX58/TDRD9 |
| GO:0070888 | GO:0070888 | E-box binding | 24 | 0.0795518 | 0.0045991 | 0.0229235 | 0.0139183 | ASCL2/MITF/NEUROD6/BHLHE41/ATOH7 |
| GO:1902936 | GO:1902936 | phosphatidylinositol bisphosphate binding | 47 | -0.1076329 | 0.0045991 | 0.0229235 | 0.0139183 | SYT1/OSBPL2/RAB35/PLCB1/PHLDA3/LAPTM4B/TTPAL/SNX21/SYT10 |
| GO:0019003 | GO:0019003 | GDP binding | 36 | -0.2405126 | 0.0047990 | 0.0235522 | 0.0143000 | RALB/RAB11B/RERG/DIRAS1 |
| GO:0051018 | GO:0051018 | protein kinase A binding | 26 | -0.1791389 | 0.0049990 | 0.0241618 | 0.0146701 | PKIA/RYR2/AKAP1/GSKIP/LRRK2/DACT2 |
| GO:0003774 | GO:0003774 | cytoskeletal motor activity | 36 | -0.1791040 | 0.0051990 | 0.0247533 | 0.0150292 | DNAH5/KIF5A/MYH7/STARD9/MYH7B/DYNC2H1/MYO19/KIFC2 |
| GO:0000287 | GO:0000287 | magnesium ion binding | 86 | 0.1174816 | 0.0059988 | 0.0273374 | 0.0165982 | ADCY2/ME1/PDE2A/SNCA/DYRK3/DYRK2/NEK6/NUDT5/MAST3/THG1L/PI4K2A/SRR |
| GO:0016811 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides | 26 | 0.1725263 | 0.0059988 | 0.0273374 | 0.0165982 | AGA/HINT1/HDAC9/NIT2 |
| GO:0031490 | GO:0031490 | chromatin DNA binding | 40 | 0.1017134 | 0.0059988 | 0.0273374 | 0.0165982 | CEBPA/HNRNPC/INSM1/SMAD3/THRA/THRB |
| GO:0019210 | GO:0019210 | kinase inhibitor activity | 38 | -0.1455030 | 0.0061988 | 0.0278508 | 0.0169099 | CDKN1B/CDKN2C/CDKN2D/PKIA/SNCA/AKT1S1 |
| GO:0004386 | GO:0004386 | helicase activity | 44 | 0.1319631 | 0.0063987 | 0.0283499 | 0.0172129 | CHD3/DDX10/RFC3/SMARCA1/DDX54/DHX58/TDRD9 |
| GO:0016407 | GO:0016407 | acetyltransferase activity | 27 | 0.1616316 | 0.0067986 | 0.0297091 | 0.0180382 | ACAT2/TAF10/NCOA3/NAA10/SPHK1 |
| GO:0003712 | GO:0003712 | transcription coregulator activity | 178 | 0.0780972 | 0.0071986 | 0.0310316 | 0.0188412 | BCL3/CRYM/FGF2/PRMT2/LMO1/PCBD1/TRIM27/SMARCD3/TLE2/UBE3A/NCOA3/ZBED1/HDAC9/BASP1/SUB1/WWC1/PHF24/TDP2/WNT4/YEATS2/LMO3/PRDM8/MTA3/SFMBT2/TRIB3/MMS19/DDX54/RIOX2/WTIP/JAZF1/MED11/HSBP1L1 |
| GO:0003924 | GO:0003924 | GTPase activity | 130 | -0.1055985 | 0.0073985 | 0.0314684 | 0.0191064 | DDX3X/EIF2S3/GNAS/RAB27B/RALB/TUBB2A/RND2/RAB11B/ENTPD4/RAB36/RGS6/TUBA1B/RRAS2/RHOBTB2/RHOQ/RASD2/ARL15/RASL11B/RERG/RAB3C/RHOV/ARL13B/ARL10/RAB37/RASL11A |
| GO:0015171 | GO:0015171 | amino acid transmembrane transporter activity | 37 | -0.1075989 | 0.0077984 | 0.0327329 | 0.0198741 | GRIK2/SLC6A6/SLC7A4/SLC38A2/SLC17A6/SLC25A22 |
| GO:0098632 | GO:0098632 | cell-cell adhesion mediator activity | 24 | -0.1717770 | 0.0079984 | 0.0331362 | 0.0201190 | CNTN1/CNTN5/DSCAML1/NEXN/EMB |
| GO:0005525 | GO:0005525 | GTP binding | 143 | -0.0940807 | 0.0083983 | 0.0334883 | 0.0203328 | EIF2S3/GNAS/INSR/MOCS1/RAB27B/RALB/TUBB2A/RND2/RAB11B/RAB36/TUBA1B/RRAS2/RHOBTB2/RHOQ/RASD2/EHD2/TUBD1/ARL15/THG1L/RASL11B/RERG/RAB3C/RHOV/ARL13B/ARL10/RAB37/RASL11A |
| GO:0016779 | GO:0016779 | nucleotidyltransferase activity | 43 | 0.1299954 | 0.0083983 | 0.0334883 | 0.0203328 | POLR2L/PRIM1/NUDT5/THG1L/POLE4/MB21D2/NMNAT3 |
| GO:0042393 | GO:0042393 | histone binding | 80 | 0.0929551 | 0.0083983 | 0.0334883 | 0.0203328 | CHD3/RBBP7/SNCA/CHAF1B/TRIM24/PRMT6/COPRS/YEATS2/SFMBT2/KDM7A/L3MBTL4/UHRF2/PHF13 |
| GO:0004860 | GO:0004860 | protein kinase inhibitor activity | 37 | -0.1417195 | 0.0087982 | 0.0342273 | 0.0207815 | CDKN1B/CDKN2C/CDKN2D/PKIA/SNCA/AKT1S1 |
| GO:0008757 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity | 57 | 0.1221933 | 0.0087982 | 0.0342273 | 0.0207815 | HNMT/PRMT2/PNMT/EMG1/BHMT2/METTL5/METTL9/PRMT6/PRDM8/TRMT2B |
| GO:0019001 | GO:0019001 | guanyl nucleotide binding | 150 | -0.0972264 | 0.0093981 | 0.0352706 | 0.0214149 | EIF2S3/GNAS/INSR/MOCS1/RAB27B/RALB/TUBB2A/RND2/RAB11B/RAB36/TUBA1B/RRAS2/RHOBTB2/RHOQ/RASD2/EHD2/TUBD1/ARL15/THG1L/RASL11B/RERG/RAB3C/RHOV/ARL13B/ARL10/RAB37/RASL11A |
| GO:0032561 | GO:0032561 | guanyl ribonucleotide binding | 150 | -0.0972264 | 0.0093981 | 0.0352706 | 0.0214149 | EIF2S3/GNAS/INSR/MOCS1/RAB27B/RALB/TUBB2A/RND2/RAB11B/RAB36/TUBA1B/RRAS2/RHOBTB2/RHOQ/RASD2/EHD2/TUBD1/ARL15/THG1L/RASL11B/RERG/RAB3C/RHOV/ARL13B/ARL10/RAB37/RASL11A |
| GO:0140313 | GO:0140313 | molecular sequestering activity | 20 | -0.1838905 | 0.0093981 | 0.0352706 | 0.0214149 | NFKBIA/PMEPA1/USP38 |
| GO:0003682 | GO:0003682 | chromatin binding | 199 | 0.0791305 | 0.0107978 | 0.0395921 | 0.0240387 | AR/CEBPA/CENPF/EGFR/GLI1/HNRNPC/INSM1/SMAD2/SMAD3/MITF/NFKB1/PRKAA2/SMARCA1/SMARCD3/THRA/THRB/NELFA/NCOR2/SMC2/VCX/PRMT6/MEIS3/PRDM8/MTA3/SFMBT2/OVOL2/ASXL3/SETD7/SARNP/JDP2/HES5/EPOP |
| GO:0070851 | GO:0070851 | growth factor receptor binding | 36 | 0.0630279 | 0.0107978 | 0.0395921 | 0.0240387 | SLA/VEGFA/VAV3/PDGFC/LINGO1 |
| GO:0005506 | GO:0005506 | iron ion binding | 38 | -0.1432670 | 0.0109978 | 0.0398670 | 0.0242057 | PHYH/PPEF1/SNCA/P3H3/CYP46A1/HAAO/KDM7A/CYP4Z1 |
| GO:0008276 | GO:0008276 | protein methyltransferase activity | 34 | 0.1636149 | 0.0111978 | 0.0399546 | 0.0242589 | PRMT2/PCMT1/FAM98A/METTL9/PRMT6/PRDM8 |
| GO:0003713 | GO:0003713 | transcription coactivator activity | 93 | 0.0655950 | 0.0113977 | 0.0399546 | 0.0242589 | BCL3/FGF2/PRMT2/LMO1/PCBD1/TRIM27/SMARCD3/UBE3A/NCOA3/ZBED1/SUB1/WWC1/LMO3/MTA3/MMS19 |
| GO:0043021 | GO:0043021 | ribonucleoprotein complex binding | 53 | 0.0842727 | 0.0113977 | 0.0399546 | 0.0242589 | DDX3X/EIF2S1/SRP19/IMPACT/TMEM223/NAA16/DAPL1/UHMK1/CPEB2 |
| GO:0097110 | GO:0097110 | scaffold protein binding | 36 | -0.1824772 | 0.0119976 | 0.0416004 | 0.0252581 | ATP2B4/GRIK2/IKBKB/ITPR2/KRT8/TCOF1/BAIAP2 |
| GO:0008094 | GO:0008094 | ATP-dependent activity, acting on DNA | 33 | 0.1116206 | 0.0123975 | 0.0425248 | 0.0258194 | CHD3/RFC3/SMARCA1/DSCC1/PIF1 |
| GO:1990841 | GO:1990841 | promoter-specific chromatin binding | 23 | 0.2135912 | 0.0127974 | 0.0434296 | 0.0263687 | SMAD3/TAF10/WDR13/PCGF1 |
| GO:0016835 | GO:0016835 | carbon-oxygen lyase activity | 28 | -0.1936351 | 0.0133973 | 0.0449868 | 0.0273142 | ACO1/CA7/CA12/OGG1/CA10/NEIL1 |
| GO:0001227 | GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | 103 | 0.0968938 | 0.0137972 | 0.0453744 | 0.0275496 | ASCL2/CREM/ENO1/INSM1/MITF/PPARD/MAFK/ZNF239/KLF8/BATF3/OVOL2/BHLHE41/MYPOP/HES5/SP5 |
| GO:0140678 | GO:0140678 | molecular function inhibitor activity | 169 | -0.0925229 | 0.0137972 | 0.0453744 | 0.0275496 | ATP2B4/CDKN1B/CDKN2C/CDKN2D/COL7A1/FKBP1A/GMFB/KNG1/LRPAP1/LY6H/NFKBIA/NKX3-1/SERPINB9/PKIA/PRNP/SCN1B/SLN/SNCA/STX1A/SLIT2/FRAT1/WFDC2/PPP1R16B/DKK3/GSKIP/SCN3B/PIF1/TAF3/AKT1S1/WFIKKN1 |
| GO:0008238 | GO:0008238 | exopeptidase activity | 27 | 0.1540201 | 0.0141972 | 0.0462132 | 0.0280588 | CTSC/CPE/AGTPBP1/SCPEP1 |
| GO:0001540 | GO:0001540 | amyloid-beta binding | 40 | -0.0880231 | 0.0163967 | 0.0528339 | 0.0320786 | EPHB2/GRIA1/INSR/LRPAP1/PRNP/ITM2A/PGRMC1/MAPK8IP2 |
| GO:0008170 | GO:0008170 | N-methyltransferase activity | 31 | 0.1389267 | 0.0165967 | 0.0529434 | 0.0321451 | HNMT/PRMT2/PNMT/METTL5/PRMT6 |
| GO:0001217 | GO:0001217 | DNA-binding transcription repressor activity | 106 | 0.0833669 | 0.0175965 | 0.0555770 | 0.0337442 | ASCL2/CREM/INSM1/SMAD3/MITF/PPARD/MAFK/ZNF239/KLF8/BATF3/OVOL2/BHLHE41/JDP2/ZNF418/MYPOP/HES5/SP5 |
| GO:0019208 | GO:0019208 | phosphatase regulator activity | 45 | -0.0723651 | 0.0177964 | 0.0556575 | 0.0337930 | ARPP19/PPP1R16B/PDP2/PPP4R4/PPP1R3B/ELFN2 |
| GO:0048306 | GO:0048306 | calcium-dependent protein binding | 36 | -0.1442816 | 0.0185963 | 0.0575943 | 0.0349690 | SYT1/TNNI3/CABP1/CPLX2/RBM22 |
| GO:0004252 | GO:0004252 | serine-type endopeptidase activity | 33 | -0.1126183 | 0.0187962 | 0.0576539 | 0.0350051 | C1S/CTSC/F12/RHBDL3/PRSS56 |
| GO:0043621 | GO:0043621 | protein self-association | 21 | 0.1615651 | 0.0189962 | 0.0577123 | 0.0350406 | ACVR2A/CTSC/MDH2/NKX3-1/RETREG1 |
| GO:0015293 | GO:0015293 | symporter activity | 48 | 0.1111128 | 0.0191962 | 0.0577696 | 0.0350754 | SLC1A4/SLC39A10/SLC25A22/SLC2A12 |
| GO:0050660 | GO:0050660 | flavin adenine dinucleotide binding | 28 | -0.1942033 | 0.0201960 | 0.0591482 | 0.0359124 | MAOB/SDHA/DUS1L/PYROXD1/AIFM3 |
| GO:0004857 | GO:0004857 | enzyme inhibitor activity | 126 | -0.0831490 | 0.0203959 | 0.0591482 | 0.0359124 | ATP2B4/CDKN1B/CDKN2C/CDKN2D/COL7A1/GMFB/KNG1/SERPINB9/PKIA/PRNP/SLN/SNCA/SLIT2/WFDC2/PPP1R16B/GSKIP/UBE2Z/PIF1/AKT1S1/ELFN2/WFIKKN1/SPINK8 |
| GO:0016829 | GO:0016829 | lyase activity | 74 | -0.1229521 | 0.0203959 | 0.0591482 | 0.0359124 | ACO1/ADCY2/ADCY7/ADCY9/AMD1/CA7/MGST2/MOCS1/OGG1/CA10/GGCT/ARMT1/TSEN2/SDSL |
| GO:0019888 | GO:0019888 | protein phosphatase regulator activity | 38 | -0.0673414 | 0.0203959 | 0.0591482 | 0.0359124 | ARPP19/PPP1R16B/PDP2/PPP4R4/PPP1R3B/ELFN2 |
| GO:0000149 | GO:0000149 | SNARE binding | 55 | -0.0565997 | 0.0205959 | 0.0591900 | 0.0359378 | GRIK2/SNCA/VAMP1/BAIAP3/SEC24D/SYT2/UNC13C |
| GO:0140297 | GO:0140297 | DNA-binding transcription factor binding | 184 | 0.0532555 | 0.0217956 | 0.0620787 | 0.0376917 | AR/ASCL2/BCL3/CEBPA/CREM/ESRRB/PRMT2/DNAJA1/LMO1/SMAD2/NFKBIA/NKX3-1/PPARD/MAPK3/RXRA/SMARCD3/USF2/WFS1/YWHAH/YWHAZ/NR4A3/NCOA3/TRIP12/NCOR2/HDAC9/ARNT2/DACT1/BEX1/HDAC8/LMO3/MMS19/DDX54/BHLHE41/RERG/UHRF2/DNAAF4 |
| GO:0015294 | GO:0015294 | solute:monoatomic cation symporter activity | 31 | 0.1583998 | 0.0229954 | 0.0649162 | 0.0394146 | SLC39A10/SLC25A22 |
| GO:0016747 | GO:0016747 | acyltransferase activity, transferring groups other than amino-acyl groups | 71 | 0.0789904 | 0.0239952 | 0.0671445 | 0.0407675 | NMT1/NCOA3/SPHK1/OSGEPL1/MBOAT7/PNPLA3/MOGAT1/MBOAT2/LCLAT1/CERS6/ZDHHC23 |
| GO:0016782 | GO:0016782 | transferase activity, transferring sulphur-containing groups | 27 | -0.0979314 | 0.0247950 | 0.0687793 | 0.0417601 | OXCT1/CHST1/NDST3/HS3ST1/LIAS/CHST15/HS6ST2 |
| GO:0001664 | GO:0001664 | G protein-coupled receptor binding | 95 | -0.1204375 | 0.0253949 | 0.0692391 | 0.0420393 | ADM/GNAS/JAK2/PENK/POMC/PRNP/CXCL12/UCHL1/WNT10B/HOMER3/ARHGEF11/CCL27/CNRIP1/REEP2/NECAB2/RSPO3 |
| GO:0020037 | GO:0020037 | heme binding | 35 | -0.0786031 | 0.0253949 | 0.0692391 | 0.0420393 | SDHD/NR1D1/ABCB6/CYP46A1/DGCR8/RSAD1/CYP4Z1 |
| GO:0042826 | GO:0042826 | histone deacetylase binding | 47 | 0.0777891 | 0.0277944 | 0.0751392 | 0.0456216 | BCL3/INSM1/NKX3-1/YWHAB/HDAC9/MTA3/BHLHE41/BRMS1L/KCTD21 |
| GO:0140104 | GO:0140104 | molecular carrier activity | 30 | -0.1238750 | 0.0309938 | 0.0830842 | 0.0504455 | PEX19/MTCH1/RANBP17/URM1/JDP2 |
| GO:0016836 | GO:0016836 | hydro-lyase activity | 22 | -0.1597426 | 0.0317936 | 0.0845181 | 0.0513161 | ACO1/CA7/CA12/CA10 |
| GO:0019894 | GO:0019894 | kinesin binding | 24 | 0.1374250 | 0.0323935 | 0.0854011 | 0.0518522 | SNCA/NEK6/MAPK8IP2 |
| GO:0043022 | GO:0043022 | ribosome binding | 32 | 0.1185089 | 0.0341932 | 0.0894067 | 0.0542842 | EIF2S1/SRP19/IMPACT/TMEM223/DAPL1/CPEB2 |
| GO:0046906 | GO:0046906 | tetrapyrrole binding | 36 | -0.0653762 | 0.0351930 | 0.0912728 | 0.0554172 | SDHD/NR1D1/ABCB6/CYP46A1/DGCR8/RSAD1/CYP4Z1 |
| GO:0005272 | GO:0005272 | sodium channel activity | 22 | -0.0955625 | 0.0357928 | 0.0920800 | 0.0559073 | HCN2/GRIK2/SCN3B |
| GO:0004714 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity | 30 | -0.0543900 | 0.0379924 | 0.0961871 | 0.0584010 | EFNB3/EGFR/FLT1/FLT3/RET/TYRO3/EPHA10 |
| GO:0008236 | GO:0008236 | serine-type peptidase activity | 41 | -0.0917318 | 0.0379924 | 0.0961871 | 0.0584010 | C1R/C1S/CTSC/F12/PCSK5/NAALAD2/RHBDL3/PRSS56 |
| GO:0005539 | GO:0005539 | glycosaminoglycan binding | 75 | -0.0872984 | 0.0387922 | 0.0974388 | 0.0591610 | ACAN/DPYSL3/EXTL2/FGF9/KNG1/LRPAP1/PRNP/PTPRF/SLIT1/VEGFA/SLIT2/ADAMTS5/FSTL1/RSPO3/RSPO2 |
| GO:0052689 | GO:0052689 | carboxylic ester hydrolase activity | 45 | -0.1022666 | 0.0421916 | 0.1051493 | 0.0638425 | BAAT/NDST3/ACOT2/ABHD12/PNPLA3/ABHD3/GDPD1 |
| GO:0051015 | GO:0051015 | actin filament binding | 82 | -0.0632865 | 0.0431914 | 0.1068065 | 0.0648487 | ADD1/EGFR/GAS2/MARCKS/MYH7/NEB/SPTB/CORO1A/COTL1/ACTR3B/MYH7B/MYO19/TLN2/NEXN |
| GO:0001618 | GO:0001618 | virus receptor activity | 25 | -0.1054682 | 0.0459908 | 0.1124798 | 0.0682933 | EFNB2/EFNB3/EGFR/IDE/ITGB1/SLC20A2 |
| GO:0008013 | GO:0008013 | beta-catenin binding | 34 | -0.0612175 | 0.0461908 | 0.1124798 | 0.0682933 | AR/CTNND2/CXADR/PXN/RORA |
| GO:0003779 | GO:0003779 | actin binding | 172 | -0.0577877 | 0.0475905 | 0.1142458 | 0.0693655 | ADD1/EGFR/EPB41/EPB41L1/GAS2/GMFB/HPCA/ITGB1/MARCKS/MYH7/NEB/SNCA/SPTB/UTRN/YWHAH/TMSB10/CORO1A/KLHL2/STK38L/COTL1/MYRIP/KLHL20/SNTG2/ACTR3B/MYH7B/CORO7/MYO19/TLN2/NEXN/PACRG |
| GO:0008234 | GO:0008234 | cysteine-type peptidase activity | 64 | 0.0685493 | 0.0477904 | 0.1142458 | 0.0693655 | CTSC/CTSO/HINT1/UCHL1/UCHL3/USP5/USP13/OTUB2/USP45 |
| GO:0005544 | GO:0005544 | calcium-dependent phospholipid binding | 31 | -0.1026135 | 0.0479904 | 0.1142458 | 0.0693655 | DOC2A/CPNE7/SYT17/SYT15/SYT3/SYT2 |
| GO:0008017 | GO:0008017 | microtubule binding | 95 | 0.1028842 | 0.0489902 | 0.1157620 | 0.0702861 | CENPF/CETN2/DYNC1I1/FGF13/GAS2/GLI1/KIF2A/KIF3C/OGG1/PAFAH1B1/PRNP/STIM1/KIF21B/HDGFL3/NAV3/JAKMIP1 |
| GO:0043177 | GO:0043177 | organic acid binding | 74 | -0.1601714 | 0.0505899 | 0.1186630 | 0.0720475 | ASS1/CRABP1/PC/PHYH/PPARD/RYR2/SCP2/GLRA3/P3H3/UBR2 |
| GO:0008146 | GO:0008146 | sulfotransferase activity | 23 | -0.0862994 | 0.0515897 | 0.1198838 | 0.0727887 | CHST1/NDST3/HS3ST1/CHST15/HS6ST2 |
| GO:0005200 | GO:0005200 | structural constituent of cytoskeleton | 48 | -0.0950032 | 0.0523895 | 0.1198838 | 0.0727887 | ANK1/ANK3/EPB41/SPTB/TUBB2A/INA/SORBS3/TUBD1/TLN2 |
| GO:0005319 | GO:0005319 | lipid transporter activity | 54 | 0.0507486 | 0.0523895 | 0.1198838 | 0.0727887 | RBP4/C2CD2L/OSBPL2/SLC27A2/ATP10D/PITPNM2/PCTP/ANO3 |
| GO:0017124 | GO:0017124 | SH3 domain binding | 51 | -0.0462300 | 0.0529894 | 0.1198838 | 0.0727887 | DPYSL3/PAK3/SH3BGRL/TOM1L1/SOCS7/ELMO2/ARHGAP27 |
| GO:0035064 | GO:0035064 | methylated histone binding | 20 | 0.1269467 | 0.0529894 | 0.1198838 | 0.0727887 | CBX6/ING3/KDM7A/PHF13 |
| GO:0038024 | GO:0038024 | cargo receptor activity | 25 | -0.1320854 | 0.0535893 | 0.1199914 | 0.0728540 | ASGR1/INSR/SORL1/TMED10/SCARA3/SCARA5 |
| GO:0140272 | GO:0140272 | exogenous protein binding | 26 | -0.0981483 | 0.0537892 | 0.1199914 | 0.0728540 | EFNB2/EFNB3/EGFR/IDE/ITGB1/SLC20A2 |
| GO:0016705 | GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | 50 | -0.0672752 | 0.0545891 | 0.1209300 | 0.0734239 | FADS3/PHYH/CYP46A1/HSPBAP1/KDM7A/TYW5/CYP4Z1/CYP4X1 |
| GO:0031406 | GO:0031406 | carboxylic acid binding | 72 | -0.1610053 | 0.0553889 | 0.1218556 | 0.0739859 | ASS1/CRABP1/PC/PHYH/PPARD/SCP2/GLRA3/P3H3/UBR2 |
| GO:0022834 | GO:0022834 | ligand-gated channel activity | 75 | -0.0867962 | 0.0557888 | 0.1218948 | 0.0740097 | HCN2/GABRA5/GABRB1/GRIK2/KCNJ14/KCNN1/TRPC3/GLRA3/KCNMB4/KCNT1 |
| GO:0141108 | GO:0141108 | transporter regulator activity | 68 | -0.0933745 | 0.0589882 | 0.1280055 | 0.0777199 | ATP2B4/FGF13/GPLD1/SCN1B/KCNAB3/KCNMB4/HPCAL4/FXYD6/SCN3B/STIM2/LRRC38/SCLT1 |
| GO:0099106 | GO:0099106 | ion channel regulator activity | 65 | -0.0884395 | 0.0593881 | 0.1280055 | 0.0777199 | ATP2B4/FGF13/GPLD1/SCN1B/YWHAH/KCNAB3/KCNMB4/HPCAL4/FXYD6/SCN3B/STIM2/LRRC38/SCLT1 |
| GO:0061629 | GO:0061629 | RNA polymerase II-specific DNA-binding transcription factor binding | 138 | 0.0573452 | 0.0627874 | 0.1344241 | 0.0816170 | AR/CEBPA/ESRRB/PRMT2/DNAJA1/SMAD2/NFKBIA/NKX3-1/RXRA/SMARCD3/YWHAH/NR4A3/NCOA3/TRIP12/NCOR2/HDAC9/ARNT2/DACT1/HES6/BEX1/MMS19/DDRGK1/DDX54/BHLHE41/RERG/UHRF2/DNAAF4 |
| GO:0016887 | GO:0016887 | ATP hydrolysis activity | 118 | 0.0672515 | 0.0645871 | 0.1372904 | 0.0833573 | ATP2B4/DDX10/DNAH5/KIF2A/KIF3C/RFC3/TAP1/HSP90B1/ABCB6/KIF21B/DDX54/DHX58/PIF1/NAV3/TDRD9/DNAH2/FIGNL2 |
| GO:0030374 | GO:0030374 | nuclear receptor coactivator activity | 21 | 0.1097426 | 0.0649870 | 0.1372904 | 0.0833573 | FGF2/NCOA3 |
| GO:0042578 | GO:0042578 | phosphoric ester hydrolase activity | 140 | 0.0363955 | 0.0657868 | 0.1375803 | 0.0835333 | DUSP3/EDNRA/EYA4/PDE1A/PDE2A/PLCB4/PPEF1/PPP1CC/PPP1R3D/PTPRA/PTPRF/PTPRR/PUDP/PLPP1/PLPPR4/BCKDK/INPP5F/NT5DC3/TDP2/PLPPR2/PLPPR3/PLCD4/PPM1M/PPM1K |
| GO:0016247 | GO:0016247 | channel regulator activity | 67 | -0.0888943 | 0.0659868 | 0.1375803 | 0.0835333 | ATP2B4/FGF13/GPLD1/SCN1B/YWHAH/KCNAB3/KCNMB4/HPCAL4/FXYD6/SCN3B/STIM2/LRRC38/SCLT1 |
| GO:0051117 | GO:0051117 | ATPase binding | 33 | -0.0632129 | 0.0675865 | 0.1400006 | 0.0850028 | ANK1/AR/EGFR/PEX19/RALB/SLN/NKD2 |
| GO:0045296 | GO:0045296 | cadherin binding | 129 | -0.0531978 | 0.0717856 | 0.1476102 | 0.0896231 | ACVR1/ADD1/ANK3/CDH7/CDH8/CDH9/CDH10/CTNND2/DDX3X/EGFR/EIF2S3/ITGB1/LDHA/YWHAB/YWHAZ/CTNNAL1/RAB11B/BAIAP2/COBLL1/DOCK9/TES/GLOD4/EIF2A |
| GO:0042277 | GO:0042277 | peptide binding | 124 | -0.0780807 | 0.0721856 | 0.1476102 | 0.0896231 | ACVR1/GALR1/IDE/INHBA/INSR/LRPAP1/MGST2/MICB/NFKBIA/OPRM1/PCSK5/PRNP/PEX19/SSTR1/SSTR2/TAP1/VIPR2/GLP2R/ITM2A/TMEM158/QRFPR/MICA |
| GO:0017171 | GO:0017171 | serine hydrolase activity | 43 | -0.0789661 | 0.0731854 | 0.1487015 | 0.0902857 | C1R/C1S/CTSC/F12/PCSK5/NAALAD2/RHBDD2/RHBDL3/PRSS56 |
| GO:0016874 | GO:0016874 | ligase activity | 39 | -0.0986510 | 0.0775845 | 0.1566421 | 0.0951069 | ASS1/ATP5F1B/ACSL4/UBA3/SLC27A2/ACSL6/CTPS2 |
| GO:0003697 | GO:0003697 | single-stranded DNA binding | 36 | 0.0571994 | 0.0815837 | 0.1636805 | 0.0993803 | SMC2/SUB1/TDP2/PCBP3/NABP1/CTC1 |
| GO:0015276 | GO:0015276 | ligand-gated monoatomic ion channel activity | 74 | -0.0803083 | 0.0861828 | 0.1718269 | 0.1043265 | HCN2/GABRA5/GABRB1/GRIK2/KCNJ14/KCNN1/TRPC3/GLRA3/KCNMB4/KCNT1 |
| GO:0004527 | GO:0004527 | exonuclease activity | 21 | 0.0961035 | 0.0887822 | 0.1752180 | 0.1063855 | DCPS/CNOT7/EXD3/DIS3L2/DCP2 |
| GO:0008201 | GO:0008201 | heparin binding | 55 | -0.0652300 | 0.0889822 | 0.1752180 | 0.1063855 | KNG1/PTPRF/SLIT1/VEGFA/SLIT2/ADAMTS5/FSTL1/RSPO3/RSPO2 |
| GO:0044325 | GO:0044325 | transmembrane transporter binding | 67 | -0.0664090 | 0.0913817 | 0.1779126 | 0.1080215 | ANK1/ANK3/FGF13/FKBP1A/KCNC1/PRNP/SCN1B/YWHAH/YWHAZ/KCNAB3/PACS1/SCN3B/LRRC38 |
| GO:0016853 | GO:0016853 | isomerase activity | 55 | -0.1081985 | 0.0915817 | 0.1779126 | 0.1080215 | FKBP1A/FKBP5/PDIA5/RPUSD2/PPIL3/ENOX1/TRUB1/TMEM86B/PGM2L1 |
| GO:0019199 | GO:0019199 | transmembrane receptor protein kinase activity | 39 | -0.0495474 | 0.0921816 | 0.1779126 | 0.1080215 | EFNB3/EGFR/FLT1/FLT3/RET/TYRO3/ACVR1C/EPHA10 |
| GO:0019207 | GO:0019207 | kinase regulator activity | 105 | -0.0512213 | 0.0925815 | 0.1779126 | 0.1080215 | CDKN1B/CDKN2C/CDKN2D/DDX3X/EGFR/FLT3/GMFB/PKIA/SNCA/VEGFA/YWHAB/TOM1L1/SOCS7/GSKIP/AKT1S1/CCNYL1/CD24 |
| GO:0016209 | GO:0016209 | antioxidant activity | 23 | -0.1168100 | 0.0933813 | 0.1783751 | 0.1083023 | GPX7/GSTT1/MGST2/SRXN1 |
| GO:0001221 | GO:0001221 | transcription coregulator binding | 47 | 0.0460191 | 0.0963807 | 0.1830086 | 0.1111156 | AR/PPARD/RORA/RXRA/SMARCD3/THRB/BAIAP2/WIZ |
| GO:0000049 | GO:0000049 | tRNA binding | 23 | 0.0464164 | 0.1005799 | 0.1894856 | 0.1150482 | EIF2S3/WDR6/THG1L/TYW5 |
| GO:0101005 | GO:0101005 | deubiquitinase activity | 41 | 0.0819657 | 0.1009798 | 0.1894856 | 0.1150482 | UCHL1/UCHL3/USP5/USP13/OTUB2/USP45 |
| GO:0003725 | GO:0003725 | double-stranded RNA binding | 26 | 0.0551016 | 0.1019796 | 0.1902427 | 0.1155078 | TUBA1B/SIDT2/SIDT1/DHX58 |
| GO:0140034 | GO:0140034 | methylation-dependent protein binding | 21 | 0.0964844 | 0.1051790 | 0.1950703 | 0.1184390 | CBX6/PHF20L1/KDM7A/PHF13 |
| GO:0140030 | GO:0140030 | modification-dependent protein binding | 58 | 0.0507884 | 0.1095781 | 0.2020544 | 0.1226794 | IDE/BRD2/VAV1/BABAM2/DZIP3/OPTN/ZBTB1/PHF20L1/YEATS2/KDM7A |
| GO:0048029 | GO:0048029 | monosaccharide binding | 23 | -0.1075522 | 0.1109778 | 0.2026612 | 0.1230479 | PHYH/P3H3/LMAN2L |
| GO:0004843 | GO:0004843 | cysteine-type deubiquitinase activity | 38 | 0.0787308 | 0.1111778 | 0.2026612 | 0.1230479 | UCHL1/UCHL3/USP5/USP13/OTUB2/USP45 |
| GO:0019783 | GO:0019783 | ubiquitin-like protein peptidase activity | 47 | 0.0680619 | 0.1119776 | 0.2029594 | 0.1232289 | HINT1/UCHL1/UCHL3/USP5/USP13/COPS4/OTUB2/USP45 |
| GO:0033218 | GO:0033218 | amide binding | 149 | -0.0693723 | 0.1139772 | 0.2054165 | 0.1247208 | ACVR1/FKBP1A/FKBP1B/FKBP5/GALR1/IDE/INHBA/INSR/LRPAP1/MGST2/MICB/NFKBIA/OPRM1/PCSK5/PRNP/PEX19/SCP2/SOAT1/SSTR1/SSTR2/TAP1/VIPR2/GLP2R/ITM2A/TMEM158/LAPTM4B/PNPLA3/MAP1LC3B/QRFPR/MICA |
| GO:0046982 | GO:0046982 | protein heterodimerization activity | 93 | 0.0388416 | 0.1199760 | 0.2150132 | 0.1305475 | ADRA1B/ADRA2C/DR1/GABBR1/ITGB1/SYT1/USF2/YWHAH/UBA3/KCNB2/ARNT2/ZBTB1/ABTB2/TENM3/RRAGD/BHLHE41/TAF3/JDP2/SYT10/CENPW |
| GO:0005125 | GO:0005125 | cytokine activity | 46 | -0.0944998 | 0.1269746 | 0.2262844 | 0.1373909 | GDF10/INHBA/LTB/CXCL12/VEGFA/WNT10B/CCL27/CMTM8 |
| GO:0098631 | GO:0098631 | cell adhesion mediator activity | 29 | -0.0715788 | 0.1323735 | 0.2345953 | 0.1424370 | CNTN1/ITGB1/ITGA11/NEXN |
| GO:0004713 | GO:0004713 | protein tyrosine kinase activity | 55 | -0.0386825 | 0.1353729 | 0.2385854 | 0.1448597 | EFNB3/EGFR/MATK/NEK1/PRKCD/MAP2K3/RET/TYRO3/DYRK3/DYRK2/DYRK4/STYK1/EPHA10 |
| GO:0015267 | GO:0015267 | channel activity | 196 | -0.0575686 | 0.1379724 | 0.2405093 | 0.1460277 | HCN2/GABRA3/GABRA5/GABRB1/GABRG1/GRIK2/KCNC1/KCNC3/KCNC4/KCNJ14/OCA2/OPRM1/SCN1A/SCN1B/SCN3A/TRPC3/GLRA3/KCNAB3/KCNB2/KCNMB4/SCN3B/KCNK12/SLC17A6/KCNT1/STIM2/ANO3/TMEM109/TRPM3/LRRC38/KCNG3/SLC26A11/TMEM150C |
| GO:0022803 | GO:0022803 | passive transmembrane transporter activity | 196 | -0.0575686 | 0.1379724 | 0.2405093 | 0.1460277 | HCN2/GABRA3/GABRA5/GABRB1/GABRG1/GRIK2/KCNC1/KCNC3/KCNC4/KCNJ14/OCA2/OPRM1/SCN1A/SCN1B/SCN3A/TRPC3/GLRA3/KCNAB3/KCNB2/KCNMB4/SCN3B/KCNK12/SLC17A6/KCNT1/STIM2/ANO3/TMEM109/TRPM3/LRRC38/KCNG3/SLC26A11/TMEM150C |
| GO:0019887 | GO:0019887 | protein kinase regulator activity | 93 | -0.0492509 | 0.1393721 | 0.2416288 | 0.1467075 | CDKN2C/CDKN2D/DDX3X/EGFR/GMFB/PKIA/SNCA/VEGFA/YWHAB/TOM1L1/GSKIP/AKT1S1/CCNYL1/CD24 |
| GO:0044389 | GO:0044389 | ubiquitin-like protein ligase binding | 128 | -0.0604206 | 0.1405719 | 0.2423915 | 0.1471706 | BID/CDKN1B/EGFR/GRIK2/DNAJA1/SMAD1/MC1R/NFKBIA/RALB/UBE2B/UBE2G1/UCHL1/YWHAZ/TNFRSF14/USP13/TUBA1B/PPARGC1A/SPART/LAPTM4B/MAP1LC3B/NKD2 |
| GO:0017018 | GO:0017018 | myosin phosphatase activity | 30 | -0.0735516 | 0.1415717 | 0.2428031 | 0.1474204 | DUSP3/PPEF1/PPM1B/PPP1CC/PPM1M |
| GO:0015291 | GO:0015291 | secondary active transmembrane transporter activity | 82 | 0.0486317 | 0.1437712 | 0.2452568 | 0.1489103 | SLC26A4/SLC6A6/SLC16A2/SLC22A5/SLC30A10/SLC39A10/SLC39A8/SLC25A23/SLC25A22/SLC4A9/SLC2A12/SLC26A11 |
| GO:0030246 | GO:0030246 | carbohydrate binding | 75 | -0.0677689 | 0.1449710 | 0.2459880 | 0.1493542 | ACAN/CANX/CNTN1/PHYH/PPP1R3D/PTX3/CLEC2B/PLA2R1/GPCPD1/CHID1/GALNT14/PPP1R3B/LMAN2L/PPP1R3F |
| GO:0030545 | GO:0030545 | signaling receptor regulator activity | 144 | -0.0501499 | 0.1517696 | 0.2554829 | 0.1551192 | ADM/EGFR/FGF13/FKBP1A/GAS6/GDF10/GMFB/GNRH1/IGF1/INHBA/KNG1/LY6H/PENK/POMC/VEGFA/WNT10B/SEMA7A/FGF18/SEMA4F/CCL27/DKK3/NRG4/ERFE/CMTM8/FNDC5 |
| GO:0002020 | GO:0002020 | protease binding | 41 | -0.0806527 | 0.1527694 | 0.2554829 | 0.1551192 | ECM1/ITGB1/MBP/SERPINB9/PRNP/RIPK2/CHL1/RHBDD2/CCBE1 |
| GO:0001228 | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | 139 | 0.0356478 | 0.1529694 | 0.2554829 | 0.1551192 | AR/ASCL2/CEBPA/KLF6/E2F3/ESRRB/ESRRG/MECOM/SMAD1/SMAD2/MAFG/MITF/MYB/NFIC/NFKB1/PBX3/POU3F2/RFX5/KLF10/NR4A3/ZBED1/ARNT2/MESP1/MEIS3/OVOL2/NEUROD6 |
| GO:0035591 | GO:0035591 | signaling adaptor activity | 43 | -0.0507200 | 0.1631674 | 0.2710958 | 0.1645987 | DDX3X/IFNAR2/PAG1/CDC42SE2/GNB4 |
| GO:0090079 | GO:0090079 | translation regulator activity, nucleic acid binding | 35 | 0.0762806 | 0.1645671 | 0.2720047 | 0.1651505 | EIF2S1/EIF2S3/EIF1AY/GSPT2/EIF5A2/CELF4/EIF2A/CPEB2 |
| GO:0072341 | GO:0072341 | modified amino acid binding | 40 | -0.0542680 | 0.1697660 | 0.2789065 | 0.1693410 | GAS6/MGST2/SYT17/GRAMD1B/SYT3/SYT2 |
| GO:0003714 | GO:0003714 | transcription corepressor activity | 74 | 0.0409263 | 0.1707658 | 0.2789065 | 0.1693410 | BCL3/CRYM/TLE2/HDAC9/TDP2/YEATS2/PRDM8/MTA3/TRIB3/RIOX2/WTIP/JAZF1/HSBP1L1 |
| GO:0043130 | GO:0043130 | ubiquitin binding | 39 | -0.0445804 | 0.1713657 | 0.2789065 | 0.1693410 | UCHL1/UCHL3/USP13/TOM1L1/GGA2/VPS36/NSFL1C/OTUB2 |
| GO:0019955 | GO:0019955 | cytokine binding | 37 | -0.1087110 | 0.1775645 | 0.2875283 | 0.1745758 | ACVR1/IFNAR2/ITGB1/TNFRSF11A/PDPN/KLHL20/WFIKKN1 |
| GO:0003735 | GO:0003735 | structural constituent of ribosome | 37 | -0.1402996 | 0.1849630 | 0.2979960 | 0.1809314 | RPL15/RPL27A/MRPL37/MRPL30/MRPL35/MRPL16/MRPS11 |
| GO:0005096 | GO:0005096 | GTPase activator activity | 122 | -0.0370724 | 0.1901620 | 0.3048325 | 0.1850823 | ARHGDIG/BCR/RASAL1/RGS6/USP6NL/GIT2/ARHGEF11/VAV3/RAB3GAP1/FAM13B/TBC1D13/TBC1D19/TBC1D24/ARHGAP9/ARHGAP28/ARHGAP33/ARHGAP27 |
| GO:0016410 | GO:0016410 | N-acyltransferase activity | 26 | 0.1034334 | 0.1921616 | 0.3064977 | 0.1860933 | NMT1/NCOA3/CERS6 |
| GO:0042562 | GO:0042562 | hormone binding | 41 | 0.0297653 | 0.1961608 | 0.3107664 | 0.1886851 | AR/CRYM/EGFR/INHBA/MC1R/THRA/THRB/GLP2R |
| GO:0015081 | GO:0015081 | sodium ion transmembrane transporter activity | 49 | 0.0873166 | 0.1973605 | 0.3107664 | 0.1886851 | HCN2/GRIK2/GRIK3/KCNK1/SCN2A/SCN3A/SCN9A/SCN3B |
| GO:0031625 | GO:0031625 | ubiquitin protein ligase binding | 123 | -0.0544213 | 0.1977604 | 0.3107664 | 0.1886851 | ACTG1/BID/CDKN1B/EGFR/GRIK2/DNAJA1/SMAD1/MC1R/NFKBIA/RALB/UBE2B/UBE2G1/UCHL1/YWHAZ/TNFRSF14/USP13/TUBA1B/PPARGC1A/SPART/LAPTM4B/MAP1LC3B/NKD2/PACRG |
| GO:0001216 | GO:0001216 | DNA-binding transcription activator activity | 141 | 0.0317252 | 0.2025595 | 0.3155135 | 0.1915673 | AR/ASCL2/CEBPA/KLF6/E2F3/E2F5/ERG/ESRRB/ESRRG/MECOM/SMAD1/SMAD2/MAFG/MITF/MYB/NFIC/NFKB1/PBX3/POU3F2/RFX5/KLF10/NR4A3/ZBED1/ARNT2/MESP1/MEIS3/OVOL2/NEUROD6 |
| GO:0033293 | GO:0033293 | monocarboxylic acid binding | 34 | -0.1185569 | 0.2027594 | 0.3155135 | 0.1915673 | CRABP1/CYP26A1/FABP7/PPARD/SCP2 |
| GO:0001222 | GO:0001222 | transcription corepressor binding | 21 | 0.0590390 | 0.2037592 | 0.3155301 | 0.1915774 | RORA/NEK6/FAM89B/WIZ |
| GO:0048018 | GO:0048018 | receptor ligand activity | 128 | -0.0444976 | 0.2063587 | 0.3161894 | 0.1919777 | ADM/FGF13/GAS6/GDF10/GMFB/GNRH1/IGF1/INHBA/KNG1/PENK/POMC/VEGFA/WNT10B/SEMA7A/FGF18/SEMA4F/CCL27/NRG4/ERFE/CMTM8/FNDC5 |
| GO:0005342 | GO:0005342 | organic acid transmembrane transporter activity | 59 | -0.0483623 | 0.2071586 | 0.3161894 | 0.1919777 | GRIK2/SLC26A4/SLC6A6/SLC7A4/SLC16A2/SLC16A6/MPC1/SLC17A6/SLC25A22 |
| GO:0046943 | GO:0046943 | carboxylic acid transmembrane transporter activity | 59 | -0.0483623 | 0.2071586 | 0.3161894 | 0.1919777 | GRIK2/SLC26A4/SLC6A6/SLC7A4/SLC16A2/SLC16A6/MPC1/SLC17A6/SLC25A22 |
| GO:0004674 | GO:0004674 | protein serine/threonine kinase activity | 157 | 0.0541950 | 0.2097580 | 0.3186325 | 0.1934611 | ACVR2A/ACVR2B/BCR/CAMK2D/EGFR/NEK1/PAK3/PRKAA2/PRKCD/PRKCG/MAPK1/MAPK3/MAP2K1/MAP2K3/MAP4K2/DYRK3/DYRK2/RIPK2/MAP3K6/STK17A/BCKDK/STK25/MAST3/TNIK/STK26/CAMK1G/UHMK1/ACVR1C/PNCK/TTBK2/STK32C |
| GO:0016798 | GO:0016798 | hydrolase activity, acting on glycosyl bonds | 26 | -0.0354040 | 0.2135573 | 0.3228662 | 0.1960316 | IL1RAP/MAN1A1/OGG1/MANEAL |
| GO:0005216 | GO:0005216 | monoatomic ion channel activity | 184 | -0.0541121 | 0.2171566 | 0.3267592 | 0.1983953 | HCN2/GABRA5/GABRB1/GABRG1/GRIK2/KCNC1/KCNC3/KCNC4/KCNJ14/OCA2/OPRM1/SCN1A/SCN1B/SCN3A/TRPC3/GLRA3/KCNAB3/KCNB2/KCNMB4/SCN3B/KCNK12/SLC17A6/KCNT1/STIM2/ANO3/TMEM109/TRPM3/LRRC38/KCNG3/SLC26A11/TMEM150C |
| GO:0015179 | GO:0015179 | L-amino acid transmembrane transporter activity | 20 | 0.0585808 | 0.2187562 | 0.3276209 | 0.1989185 | SLC25A22 |
| GO:0106310 | GO:0106310 | protein serine kinase activity | 132 | 0.0607317 | 0.2461508 | 0.3669257 | 0.2227828 | BCR/CAMK2D/NEK1/PAK3/PRKAA2/PRKCD/PRKCG/MAPK1/MAPK3/MAP2K1/MAP2K3/MAP4K2/DYRK3/DYRK2/RIPK2/MAP3K6/STK17A/BCKDK/STK25/MAST3/TNIK/STK26/CAMK1G/UHMK1/PNCK/TTBK2/STK32C |
| GO:0140767 | GO:0140767 | enzyme-substrate adaptor activity | 24 | 0.0725691 | 0.2489502 | 0.3693726 | 0.2242685 | SH3BGRL/KLHL2/FBXL2/FZR1 |
| GO:0005245 | GO:0005245 | voltage-gated calcium channel activity | 25 | -0.0545068 | 0.2527495 | 0.3716046 | 0.2256236 | OPRM1/CACNA1H/CACNA2D2/CACNG3/CACNG8 |
| GO:0030546 | GO:0030546 | signaling receptor activator activity | 131 | -0.0389033 | 0.2533493 | 0.3716046 | 0.2256236 | ADM/EGFR/FGF13/GAS6/GDF10/GMFB/GNRH1/IGF1/INHBA/KNG1/PENK/POMC/VEGFA/WNT10B/SEMA7A/FGF18/SEMA4F/CCL27/PDGFC/ERFE/CMTM8/FNDC5 |
| GO:0005249 | GO:0005249 | voltage-gated potassium channel activity | 57 | 0.0969053 | 0.2539492 | 0.3716046 | 0.2256236 | HCN2/KCNA4/KCNA5/KCNC4/KCNG1/KCNJ12/KCNK1/KCNK2/KCNAB3/KCNB2/KCNG3 |
| GO:0019842 | GO:0019842 | vitamin binding | 52 | -0.0527847 | 0.2567487 | 0.3739855 | 0.2270692 | CALB1/CRABP1/OGDH/PHYH/RBP4/P4HA2/P3H3/PNPO/SRR/SDSL |
| GO:0008514 | GO:0008514 | organic anion transmembrane transporter activity | 83 | -0.0354804 | 0.2623475 | 0.3804039 | 0.2309662 | GRIK2/GRIK3/SLC26A4/SLC6A6/SLC7A4/SLC16A2/SLC16A6/MPC1/SLC17A6/SLC39A8/SLC25A22/SLC4A9/SLC26A11 |
| GO:0022853 | GO:0022853 | active monoatomic ion transmembrane transporter activity | 80 | 0.0640060 | 0.2655469 | 0.3821488 | 0.2320257 | ATP2B4/ATP6V1C1/SLC26A4/SLC30A10/SLC39A10/SLC39A8/SLC25A22/SLC4A9/SLC26A11 |
| GO:0046915 | GO:0046915 | transition metal ion transmembrane transporter activity | 20 | 0.0411899 | 0.2659468 | 0.3821488 | 0.2320257 | SLC25A37/SLC30A10/SLC39A10/SLC39A8 |
| GO:1901681 | GO:1901681 | sulfur compound binding | 89 | -0.0338855 | 0.2721456 | 0.3893024 | 0.2363691 | DPYSL3/KNG1/MOCS1/PTPRF/SLIT1/SOAT1/VEGFA/ADAMTS5/FSTL1/CHST15/METTL17/PNPLA3/RSPO3/RSPO2 |
| GO:0004175 | GO:0004175 | endopeptidase activity | 103 | -0.0364315 | 0.2787443 | 0.3969617 | 0.2410195 | C1R/C1S/CTSC/F12/IDE/PCSK5/KLK10/UCHL1/USP13/ADAMTS5/RHBDD2/SPPL3/RHBDL3/ADAMTS19/PRSS56 |
| GO:0061135 | GO:0061135 | endopeptidase regulator activity | 41 | -0.0364904 | 0.2805439 | 0.3977489 | 0.2414974 | COL7A1/ANOS1/KNG1/SERPINB9/SERPINI1/PRNP/WFDC2/WFDC1/UBE2Z/WFIKKN1 |
| GO:0030276 | GO:0030276 | clathrin binding | 24 | 0.0494653 | 0.2853429 | 0.4027628 | 0.2445417 | TOM1L1/SYT17/SYT2/SCLT1/SYT10 |
| GO:0016922 | GO:0016922 | nuclear receptor binding | 58 | 0.0282455 | 0.2891422 | 0.4063275 | 0.2467060 | PRMT2/NKX3-1/RXRA/SMARCD3/YWHAH/NR4A3/NCOA3/TRIP12/NCOR2/RERG/DNAAF4 |
| GO:0030971 | GO:0030971 | receptor tyrosine kinase binding | 35 | -0.0292425 | 0.2939412 | 0.4112599 | 0.2497008 | DUSP3/GAS6/GRB14/TIAM1/PITPNM2/ELMO2/FIZ1 |
| GO:0016614 | GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | 49 | -0.0658883 | 0.2955409 | 0.4116923 | 0.2499633 | HMGCR/HSD11B1/LDHA/KCNAB3/HIBADH/HSD17B11/DCXR |
| GO:0015079 | GO:0015079 | potassium ion transmembrane transporter activity | 81 | 0.0803545 | 0.2979404 | 0.4132304 | 0.2508972 | HCN2/GRIK2/GRIK3/KCNA4/KCNA5/KCNC4/KCNG1/KCNJ12/KCNK1/KCNK2/KCNAB3/KCNB2/KCNMB2/KCNMB4/KCNG3 |
| GO:0008135 | GO:0008135 | translation factor activity, RNA binding | 27 | 0.0700502 | 0.3055389 | 0.4219347 | 0.2561821 | EIF2S1/EIF2S3/EIF1AY/GSPT2/EIF5A2/EIF2A/CPEB2 |
| GO:0030544 | GO:0030544 | Hsp70 protein binding | 24 | -0.0849682 | 0.3109378 | 0.4274290 | 0.2595180 | CDKN1B/DNAJA1/SNCA/HDAC8 |
| GO:0030170 | GO:0030170 | pyridoxal phosphate binding | 22 | -0.0459131 | 0.3135373 | 0.4274290 | 0.2595180 | PYGL/PNPO/SRR/SDSL |
| GO:0070279 | GO:0070279 | vitamin B6 binding | 22 | -0.0459131 | 0.3135373 | 0.4274290 | 0.2595180 | PYGL/PNPO/SRR/SDSL |
| GO:0015631 | GO:0015631 | tubulin binding | 137 | 0.0425610 | 0.3175365 | 0.4310389 | 0.2617098 | CETN2/DYNC1I1/FGF13/GAS2/KIF2A/KIF3C/OGG1/PRNP/SNCA/TBCE/ARHGEF7/NISCH/KIF21B/AGTPBP1/HDGFL3/STMN3/TRIM36/RCC2/MAP1LC3B/NAV3/PACRG/TTBK2/JAKMIP1/FAM110C |
| GO:0030331 | GO:0030331 | nuclear estrogen receptor binding | 20 | 0.0407471 | 0.3205359 | 0.4332667 | 0.2630625 | PRMT2/NKX3-1/DNAAF4 |
| GO:0046873 | GO:0046873 | metal ion transmembrane transporter activity | 181 | 0.0573697 | 0.3225355 | 0.4341301 | 0.2635866 | ATP2B4/HCN2/FKBP1B/GPM6A/GRIK2/GRIK3/KCNA5/KCNC4/KCNG1/KCNJ12/KCNK1/KCNK2/OPRM1/SCN3A/TRPC3/TUSC3/CACNA1H/KCNAB3/KCNB2/KCNMB2/CACNG3/KCNMB4/TRPV2/SLC30A10/SCN3B/KCNK12/TRPM3/GRIN3A/KCNG3 |
| GO:0051082 | GO:0051082 | unfolded protein binding | 38 | -0.0698944 | 0.3277345 | 0.4392743 | 0.2667100 | CANX/CLGN/DNAJA1/HSPB2/DNAJC4/HSPB6/SRSF12 |
| GO:0016298 | GO:0016298 | lipase activity | 44 | 0.0319845 | 0.3299340 | 0.4403722 | 0.2673766 | EDNRA/GPLD1/PLCB2/PLCB4/ABHD12/PNPLA3/PLCD4/PLB1 |
| GO:0005085 | GO:0005085 | guanyl-nucleotide exchange factor activity | 106 | -0.0221448 | 0.3517297 | 0.4675073 | 0.2838520 | BCR/HPS1/MCF2/RASGRF1/RPGR/TIAM1/ARHGEF11/VAV3/RAB3GAP1/IQSEC2/MCF2L/DOCK9/PSD3/THG1L/RCC2/ARFGEF3/DENND1A/ARHGEF28/PLCD4/KNDC1/DENND1B/RASGEF1C |
| GO:0016616 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | 45 | -0.0643311 | 0.3571286 | 0.4727138 | 0.2870131 | HMGCR/HSD11B1/LDHA/KCNAB3/HIBADH/HSD17B11/DCXR |
| GO:0048156 | GO:0048156 | tau protein binding | 20 | -0.0633281 | 0.3653269 | 0.4815673 | 0.2923887 | PRKAA1/SNCA/BRSK1/TTBK2 |
| GO:1990782 | GO:1990782 | protein tyrosine kinase binding | 47 | -0.0252863 | 0.3675265 | 0.4824731 | 0.2929386 | DUSP3/GAS6/GRB14/ITGB1/MFAP3/TIAM1/PITPNM2/ELMO2/FIZ1 |
| GO:0005179 | GO:0005179 | hormone activity | 36 | -0.0371345 | 0.3705259 | 0.4844171 | 0.2941189 | ADM/INHBA/KNG1/PENK/POMC/ERFE/FNDC5 |
| GO:0022804 | GO:0022804 | active transmembrane transporter activity | 142 | 0.0427638 | 0.3723255 | 0.4847830 | 0.2943411 | ATP2B4/ATP6V1C1/NDUFC2/SLC26A4/SLC6A6/SLC16A2/SLC22A5/TAP1/ABCB6/SLC30A10/SLC39A10/SLC39A8/SLC25A23/SLC25A22/SLC4A9/SLC2A12/SLC26A11 |
| GO:0004721 | GO:0004721 | phosphoprotein phosphatase activity | 70 | -0.0252976 | 0.3803239 | 0.4931843 | 0.2994420 | DUSP3/EYA4/PPEF1/PPP1CC/PPP1R3D/PTPRA/PTPRF/BCKDK/PPP1R3B/PPM1M |
| GO:0015078 | GO:0015078 | proton transmembrane transporter activity | 43 | 0.0582571 | 0.3861228 | 0.4976948 | 0.3021807 | ATP6V1C1/ATP6V1H/SLC45A4/SLC25A22 |
| GO:0005504 | GO:0005504 | fatty acid binding | 21 | -0.0785028 | 0.3869226 | 0.4976948 | 0.3021807 | CRABP1/FABP7/PPARD/SCP2 |
| GO:0004620 | GO:0004620 | phospholipase activity | 39 | 0.0292932 | 0.4091182 | 0.5241313 | 0.3182319 | EDNRA/GPLD1/PLCB2/PLCB4/ABHD12/PNPLA3/PLCD4/PLB1 |
| GO:0019902 | GO:0019902 | phosphatase binding | 77 | -0.0180656 | 0.4147171 | 0.5291790 | 0.3212966 | ANK1/ATP2B4/CTSC/EGFR/PPP1CC/PPP1R3D/SPHK1/DZIP3/FBXL2/PPP1R3F/GRIN3A |
| GO:0022843 | GO:0022843 | voltage-gated monoatomic cation channel activity | 84 | 0.0560223 | 0.4193161 | 0.5329157 | 0.3235654 | HCN2/KCNA4/KCNA5/KCNC4/KCNG1/KCNJ12/KCNK1/KCNK2/OPRM1/CACNA1H/KCNAB3/KCNB2/CACNG3/KCNK12/KCNG3 |
| GO:0003823 | GO:0003823 | antigen binding | 20 | 0.0709302 | 0.4253149 | 0.5383947 | 0.3268920 | LAG3/TAP1/SPON2 |
| GO:0005548 | GO:0005548 | phospholipid transporter activity | 24 | 0.0254142 | 0.4319136 | 0.5445867 | 0.3306516 | C2CD2L/PITPNM2/PCTP/ANO3 |
| GO:0016763 | GO:0016763 | pentosyltransferase activity | 25 | -0.0190157 | 0.4365127 | 0.5482187 | 0.3328568 | MTAP/LARGE1/XYLT1/XYLT2 |
| GO:0030159 | GO:0030159 | signaling receptor complex adaptor activity | 22 | -0.0275338 | 0.4397121 | 0.5500712 | 0.3339815 | GNB2/GNB3/SORBS1/GNB4/MMS19 |
| GO:0016810 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | 47 | 0.0169119 | 0.4497101 | 0.5603809 | 0.3402412 | CRMP1/DPYSL3/GDA/HDAC9/DPYSL4/NGLY1/HDAC8/NIT2/MBLAC2 |
| GO:0016504 | GO:0016504 | peptidase activator activity | 22 | 0.0382988 | 0.4757049 | 0.5904663 | 0.3585079 | CTSC/NKX3-1/PSME2 |
| GO:0008083 | GO:0008083 | growth factor activity | 43 | -0.0335550 | 0.4905019 | 0.6064733 | 0.3682266 | FGF13/GDF10/GMFB/INHBA/VEGFA |
| GO:0051020 | GO:0051020 | GTPase binding | 144 | -0.0197708 | 0.5188962 | 0.6373836 | 0.3869942 | DAPK3/MEGF9/GNB2/GRIA1/PAK3/TRIP11/USP6NL/DOCK4/GIT2/NCKAP1/RAB3GAP1/RIMS1/GGA2/MYRIP/ANKFY1/RAPGEF6/TBC1D13/FNBP1L/CCDC186/TMEM127/RADIL/RCC2/CDC42SE2/RRAGD/DENND1B/RILPL2 |
| GO:0005244 | GO:0005244 | voltage-gated monoatomic ion channel activity | 94 | 0.0433434 | 0.5214957 | 0.6373836 | 0.3869942 | HCN2/KCNA5/KCNC4/KCNG1/KCNJ12/KCNK1/KCNK2/OPRM1/CACNA1H/KCNAB3/KCNB2/CACNG3/KCNMB4/KCNK12/GRIN3A/LRRC38/KCNG3 |
| GO:0022832 | GO:0022832 | voltage-gated channel activity | 94 | 0.0433434 | 0.5214957 | 0.6373836 | 0.3869942 | HCN2/KCNA5/KCNC4/KCNG1/KCNJ12/KCNK1/KCNK2/OPRM1/CACNA1H/KCNAB3/KCNB2/CACNG3/KCNMB4/KCNK12/GRIN3A/LRRC38/KCNG3 |
| GO:0046332 | GO:0046332 | SMAD binding | 32 | -0.0239137 | 0.5428914 | 0.6610014 | 0.4013340 | ACVR1/FKBP1A/SMAD1/SMAD2 |
| GO:0016651 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H | 28 | -0.0567335 | 0.5456909 | 0.6618836 | 0.4018696 | NDUFC2/DCXR/ENOX1/RNLS/AIFM3 |
| GO:0005543 | GO:0005543 | phospholipid binding | 196 | -0.0149102 | 0.5480904 | 0.6622759 | 0.4021078 | EPB41/GAP43/GAS6/SNCA/SYT1/RASAL1/DOC2B/DOC2A/BAIAP3/CPNE6/C2CD2L/OSBPL2/TOM1L1/RAB35/NISCH/MCF2L/CPNE7/VPS36/SNX7/RAPGEF6/SYT17/EPN3/LAPTM4B/GRAMD1B/PITPNM2/DENND1A/PCTP/TTPAL/PNPLA3/MAP1LC3B/SYT3/ZFYVE19/ARHGAP33/SYT2/DENND1B/SYT10/UNC13C |
| GO:0005178 | GO:0005178 | integrin binding | 73 | -0.0255683 | 0.5656869 | 0.6809589 | 0.4134514 | CXADR/EGFR/IGF1/ITGB1/CXCL12/ICAM5/UTRN/SEMA7A/ADAMTS5/NISCH/ITGA11/TLN2 |
| GO:0140375 | GO:0140375 | immune receptor activity | 37 | -0.0522264 | 0.5782843 | 0.6935064 | 0.4210697 | CMKLR1/FLT3/GFRA1/IFNAR2/IL1RAP/IL12RB2/IL17RA/IFNLR1 |
| GO:0016655 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor | 20 | -0.0661059 | 0.5914817 | 0.7066766 | 0.4290662 | NDUFA4/NDUFC2/DCXR/AIFM2 |
| GO:0015399 | GO:0015399 | primary active transmembrane transporter activity | 59 | 0.0438318 | 0.5940812 | 0.7071339 | 0.4293438 | ATP2B4/ATP6V1C1/NDUFC2/TAP1/ABCB6 |
| GO:0001786 | GO:0001786 | phosphatidylserine binding | 29 | 0.0189744 | 0.5994801 | 0.7085109 | 0.4301799 | GAP43/GAS6/SYT1/CPNE6/SYT17/SYT2/SYT10 |
| GO:0017046 | GO:0017046 | peptide hormone binding | 20 | 0.0192025 | 0.5996801 | 0.7085109 | 0.4301799 | INHBA/GLP2R |
| GO:0005267 | GO:0005267 | potassium channel activity | 69 | 0.0399365 | 0.6108778 | 0.7190776 | 0.4365955 | HCN2/GRIK2/GRIK3/KCNA5/KCNC4/KCNG1/KCNJ12/KCNAB3/KCNB2/KCNMB4/KCNK12/KCNG3 |
| GO:0030295 | GO:0030295 | protein kinase activator activity | 47 | 0.0187463 | 0.6452709 | 0.7567700 | 0.4594809 | DDX3X/EGFR/VEGFA/TOM1L1/MOB1B/CCNYL1/CD24 |
| GO:0019905 | GO:0019905 | syntaxin binding | 36 | 0.0153209 | 0.6522695 | 0.7621758 | 0.4627630 | VAMP1/BAIAP3/TMED10/SYT17/SYT2/TXLNB/UNC13C |
| GO:0019838 | GO:0019838 | growth factor binding | 50 | -0.0125865 | 0.6638672 | 0.7707818 | 0.4679883 | EGFR/FLT1/FLT3/IGFALS/IGFBP5/KAZALD1/NKD2/ACVR1C |
| GO:0051087 | GO:0051087 | protein-folding chaperone binding | 53 | -0.0275880 | 0.6644671 | 0.7707818 | 0.4679883 | CDKN1B/CTSC/DNAJA1/PRNP/SNCA/TBCE/USP13/PDPN/HDAC8/FNIP2/PACRG |
| GO:0140657 | GO:0140657 | ATP-dependent activity | 176 | 0.0139705 | 0.6790642 | 0.7829481 | 0.4753752 | ATP2B4/ATP6V1C1/DNAH5/ACSL4/KIF2A/KIF3C/MYH7/RECQL/RFC3/TAP1/HSP90B1/UBA3/ABCB6/SMC2/SLC27A2/KIF21B/ATP10D/MYH7B/DDX31/DSCC1/DHX58/PIF1/NAV3/TDRD9/DNAH14/DNAH2/FIGNL2 |
| GO:0042626 | GO:0042626 | ATPase-coupled transmembrane transporter activity | 41 | 0.0301969 | 0.6798640 | 0.7829481 | 0.4753752 | ATP2B4/ATP6V1C1/TAP1/ABCB6 |
| GO:0016755 | GO:0016755 | aminoacyltransferase activity | 167 | 0.0143472 | 0.6876625 | 0.7890803 | 0.4790984 | BIRC3/TRIM27/RING1/RPGR/TGM1/UBE2B/UBE2D2/UBE2E2/UBE2G1/RNF113A/UBA3/ZBED1/TRIP12/DZIP3/NEDD4L/RNF11/KLHL20/TRIM36/PELI1/RNF150/SH3RF1/CCNB1IP1/UBE2O/PJA1/UBE2Z/UHRF2/HECTD2/MED11 |
| GO:0005518 | GO:0005518 | collagen binding | 28 | -0.0270854 | 0.6916617 | 0.7900506 | 0.4796875 | ITGB1/TGFBI/ITGA11/CCBE1/RELL2 |
| GO:0022836 | GO:0022836 | gated channel activity | 139 | -0.0203717 | 0.6934613 | 0.7900506 | 0.4796875 | HCN2/FKBP1B/GABRA3/GABRA5/GABRB1/GABRB3/GABRG1/GRIK2/GRIK3/KCNC3/KCNC4/KCNG1/KCNJ14/OPRM1/TRPC3/GLRA3/KCNAB3/KCNB2/KCNMB4/KCNK12/KCNT1/TMEM109/LRRC38/KCNG3/TMEM150C |
| GO:0120013 | GO:0120013 | lipid transfer activity | 25 | 0.0153574 | 0.7018596 | 0.7953048 | 0.4828777 | C2CD2L/OSBPL2/GRAMD1B/PITPNM2/STARD5 |
| GO:0035254 | GO:0035254 | glutamate receptor binding | 27 | -0.0144326 | 0.7030594 | 0.7953048 | 0.4828777 | GNAS/PRNP/HOMER3/NECAB2 |
| GO:0005516 | GO:0005516 | calmodulin binding | 104 | -0.0123271 | 0.7056589 | 0.7954247 | 0.4829505 | ADD1/ATP2B2/ATP2B4/EGFR/EPB41/MARCKS/MYH7/PCNT/PCP4/PDE1A/SNTB1/SYT1/SPHK1/FBXL2/CAMK1G/ITPKC/PNCK/UNC13C |
| GO:0015297 | GO:0015297 | antiporter activity | 39 | -0.0068023 | 0.7082583 | 0.7955437 | 0.4830227 | SLC26A4/SLC9A5/SLC30A10/SLC17A6/SLC25A23/SLC4A9/SLC26A11 |
| GO:0017080 | GO:0017080 | sodium channel regulator activity | 22 | -0.0201764 | 0.7150570 | 0.8003620 | 0.4859482 | ATP2B4/FGF13/FXYD6/SCN3B/SCLT1 |
| GO:0061134 | GO:0061134 | peptidase regulator activity | 55 | 0.0109262 | 0.7254549 | 0.8091612 | 0.4912908 | CTSC/ANOS1/KNG1/NKX3-1/SERPINB9/SERPINI1/PRNP/WFDC2/WFDC1/UBE2Z |
| GO:0005261 | GO:0005261 | monoatomic cation channel activity | 144 | 0.0175413 | 0.7388522 | 0.8210393 | 0.4985026 | HCN2/FKBP1B/GPM6A/GRIK2/GRIK3/KCNA5/KCNC4/KCNG1/KCNJ12/OPRM1/SCN3A/TRPC3/KCNAB3/KCNB2/KCNMB4/SCN3B/KCNK12/TRPM3/GRIN3A/LRRC38/KCNG3 |
| GO:0051287 | GO:0051287 | NAD binding | 26 | -0.0155652 | 0.7412517 | 0.8210393 | 0.4985026 | ALDH1A3/HADH/HIBADH/RNLS |
| GO:0004197 | GO:0004197 | cysteine-type endopeptidase activity | 34 | 0.0097224 | 0.7488502 | 0.8265855 | 0.5018701 | CTSC/CTSO/UCHL1/USP13 |
| GO:0061630 | GO:0061630 | ubiquitin protein ligase activity | 117 | -0.0106526 | 0.7538492 | 0.8292342 | 0.5034783 | BIRC3/TRIM27/RING1/RPGR/RNF113A/TRIP12/DZIP3/NEDD4L/RNF11/PELI1/RNF150/SH3RF1/CCNB1IP1/UBE2O/PJA1/LONRF3/UHRF2/HECTD2/MED11 |
| GO:0031267 | GO:0031267 | small GTPase binding | 127 | -0.0104864 | 0.7646471 | 0.8326821 | 0.5055717 | DAPK3/DIAPH2/MEGF9/GRIA1/PAK3/TRIP11/USP6NL/DOCK4/GIT2/ABI2/NCKAP1/RAB3GAP1/RIMS1/GGA2/MYRIP/ANKFY1/RAPGEF6/TBC1D13/CCDC186/TMEM127/BRK1/RCC2/CDC42SE2/DENND1B/RILPL2 |
| GO:0009055 | GO:0009055 | electron transfer activity | 42 | -0.0296718 | 0.7652470 | 0.8326821 | 0.5055717 | COX7A1/MAOB/NDUFC2/SDHD/P4HA2/HAAO/AIFM2 |
| GO:0045182 | GO:0045182 | translation regulator activity | 45 | 0.0131264 | 0.7660468 | 0.8326821 | 0.5055717 | EIF2S3/IGF2BP3/GSPT2/EIF5A2/CELF4/EIF2A/DAPL1/CPEB2/NANOS1 |
| GO:0005201 | GO:0005201 | extracellular matrix structural constituent | 53 | -0.0077437 | 0.7714457 | 0.8326821 | 0.5055717 | ACAN/COL7A1/COL12A1/ANOS1/LAMA3/LAMA4/TGFBI/THSD4/COL21A1/LAMA1 |
| GO:0004866 | GO:0004866 | endopeptidase inhibitor activity | 33 | -0.0073221 | 0.7726455 | 0.8326821 | 0.5055717 | ANOS1/KNG1/SERPINB9/SERPINI1/PRNP/WFDC2/WFDC1/UBE2Z |
| GO:0030414 | GO:0030414 | peptidase inhibitor activity | 33 | -0.0073221 | 0.7726455 | 0.8326821 | 0.5055717 | ANOS1/KNG1/SERPINB9/SERPINI1/PRNP/WFDC2/WFDC1/UBE2Z |
| GO:0032182 | GO:0032182 | ubiquitin-like protein binding | 45 | -0.0089558 | 0.7960408 | 0.8550068 | 0.5191264 | UCHL1/UCHL3/USP13/TOM1L1/VPS36/DCUN1D2/NSFL1C/OTUB2 |
| GO:0016791 | GO:0016791 | phosphatase activity | 104 | 0.0058036 | 0.7998400 | 0.8562046 | 0.5198536 | DUSP3/EYA4/PPEF1/PPP1CC/PPP1R3D/PTPRA/PTPRF/PUDP/PLPP1/PLPPR4/BCKDK/INPP5F/NT5DC3/NT5M/PLPPR2/PLPPR3/PPM1M |
| GO:0019787 | GO:0019787 | ubiquitin-like protein transferase activity | 165 | 0.0087951 | 0.8046391 | 0.8584611 | 0.5212237 | BIRC3/TRIM27/RING1/RPGR/UBE2B/UBE2D2/UBE2E2/UBE2G1/RNF113A/UBA3/ZBED1/TRIP12/DZIP3/NEDD4L/RNF11/KLHL20/TRIM36/PELI1/RNF150/SH3RF1/CCNB1IP1/UBE2O/PJA1/UBE2Z/UHRF2/HECTD2/MED11 |
| GO:0061659 | GO:0061659 | ubiquitin-like protein ligase activity | 124 | -0.0081285 | 0.8136373 | 0.8651676 | 0.5252956 | BIRC3/TRIM27/RING1/RPGR/UBE2D2/RNF113A/ZBED1/TRIP12/DZIP3/NEDD4L/RNF11/PELI1/RNF150/SH3RF1/CCNB1IP1/UBE2O/PJA1/LONRF3/RNF157/UHRF2/HECTD2/MED11 |
| GO:0016765 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups | 20 | -0.0121253 | 0.8192362 | 0.8655633 | 0.5255359 | GSTT1/HMBS/NUS1 |
| GO:0030145 | GO:0030145 | manganese ion binding | 28 | 0.0053554 | 0.8194361 | 0.8655633 | 0.5255359 | ADCY2/PPEF1/DYRK2/LARGE1/PPM1M |
| GO:0019829 | GO:0019829 | ATPase-coupled monoatomic cation transmembrane transporter activity | 28 | -0.0195224 | 0.8252350 | 0.8688117 | 0.5275082 | ATP2B2/ATP2B4/ATP6V1C1 |
| GO:0031593 | GO:0031593 | polyubiquitin modification-dependent protein binding | 22 | -0.0111566 | 0.8292342 | 0.8701503 | 0.5283209 | BABAM2/DZIP3/ZBTB1 |
| GO:0004896 | GO:0004896 | cytokine receptor activity | 29 | -0.0151918 | 0.8386323 | 0.8771269 | 0.5325568 | FLT3/IFNAR2/IL1RAP/IL12RB2/IL17RA/IFNLR1 |
| GO:0008233 | GO:0008233 | peptidase activity | 164 | -0.0062599 | 0.8596281 | 0.8961482 | 0.5441058 | C1R/C1S/CTSC/CPE/F12/IDE/CHMP1A/PCSK5/RELN/KLK10/UCHL1/UCHL3/USP13/NAALAD2/ADAMTS5/CFAP44/RHBDD2/DPP10/SCPEP1/OTUB2/ERMP1/MINDY3/USP45/SPPL3/TRABD2A/RHBDL3/ADAMTS19/PRSS56 |
| GO:0004722 | GO:0004722 | protein serine/threonine phosphatase activity | 38 | -0.0082509 | 0.8648270 | 0.8986313 | 0.5456135 | DUSP3/PPEF1/PPP1CC/PPP1R3D/BCKDK/PPM1M |
| GO:0004842 | GO:0004842 | ubiquitin-protein transferase activity | 151 | -0.0055849 | 0.8722256 | 0.9033765 | 0.5484945 | BIRC3/TRIM27/RING1/RPGR/UBE2B/UBE2D2/UBE2E2/UBE2G1/RNF113A/TRIP12/DZIP3/NEDD4L/RNF11/KLHL20/TRIM36/PELI1/RNF150/SH3RF1/CCNB1IP1/UBE2O/PJA1/UBE2Z/LONRF3/UHRF2/HECTD2/MED11 |
| GO:0008237 | GO:0008237 | metallopeptidase activity | 49 | -0.0091120 | 0.8834233 | 0.9092769 | 0.5520770 | CPE/IDE/CHMP1A/KLK7/NAALAD2/ADAMTS5/ERMP1/TRABD2A/ADAMTS19 |
| GO:0004222 | GO:0004222 | metalloendopeptidase activity | 31 | -0.0087407 | 0.8836233 | 0.9092769 | 0.5520770 | IDE/KLK7/ADAMTS5/TRABD2A/ADAMTS19 |
| GO:0019209 | GO:0019209 | kinase activator activity | 50 | 0.0053237 | 0.8888222 | 0.9116858 | 0.5535397 | DDX3X/EGFR/VEGFA/TOM1L1/MOB1B/CCNYL1/CD24 |
| GO:0099094 | GO:0099094 | ligand-gated monoatomic cation channel activity | 52 | 0.0078788 | 0.8986203 | 0.9187816 | 0.5578479 | HCN2/FKBP1B/GRIK2/GRIK3/KCNJ12/TRPC3/KCNMB4/KCNT1 |
| GO:0005262 | GO:0005262 | calcium channel activity | 49 | 0.0057379 | 0.9034193 | 0.9207373 | 0.5590353 | FKBP1B/GPM6A/OPRM1/TRPC3/CACNA1H/CACNG3/TRPV2/STIM2/TRPM3/GRIN3A |
| GO:1901981 | GO:1901981 | phosphatidylinositol phosphate binding | 73 | -0.0024552 | 0.9230154 | 0.9377131 | 0.5693424 | GAP43/SYT1/OSBPL2/RAB35/VPS36/LAPTM4B/DENND1A/TTPAL/ZFYVE19/DENND1B/SYT10 |
| GO:0005126 | GO:0005126 | cytokine receptor binding | 59 | 0.0022422 | 0.9392122 | 0.9511387 | 0.5774939 | FKBP1A/IL1RAP/LTB/SMAD2/CXCL12/VEGFA/BABAM2/CCL27/RASL11B/IL34 |
| GO:0019903 | GO:0019903 | protein phosphatase binding | 60 | -0.0017380 | 0.9432114 | 0.9521659 | 0.5781176 | ANK1/ATP2B4/EGFR/PPP1CC/PPP1R3D/SPHK1/FBXL2/PPP1R3F/GRIN3A |
| GO:0015085 | GO:0015085 | calcium ion transmembrane transporter activity | 59 | -0.0026411 | 0.9534093 | 0.9594245 | 0.5825247 | ATP2B4/FKBP1B/GPM6A/OPRM1/TRPC3/SLC30A10/STIM2/TRPM3/GRIN3A |
| GO:0035091 | GO:0035091 | phosphatidylinositol binding | 112 | 0.0001203 | 0.9962008 | 0.9993335 | 0.6067558 | EPB41/GAP43/SYT1/C2CD2L/OSBPL2/TOM1L1/RAB35/NISCH/MCF2L/VPS36/SNX7/LAPTM4B/PITPNM2/DENND1A/TTPAL/ZFYVE19/ARHGAP33/DENND1B/SYT10 |
| GO:0031072 | GO:0031072 | heat shock protein binding | 44 | 0.0000137 | 1.0000000 | 1.0000000 | 0.6071605 | FKBP5/DNAJA1/SNCA/TFAM/HDAC8/PACRG |