 BrainEnrich is an R package designed to facilitate the integration of brain imaging data with transcriptomic profiles. It enables researchers to explore the molecular underpinnings of brain phenotypes by performing enrichment analysis of predefined gene sets. Whether working at the group or individual level, the package offers a flexible and powerful tool for examining associations between brain imaging phenotypes (e.g., cortical thickness) and gene expression, using a variety of statistical models, null models, and aggregation methods.
BrainEnrich is an R package designed to facilitate the integration of brain imaging data with transcriptomic profiles. It enables researchers to explore the molecular underpinnings of brain phenotypes by performing enrichment analysis of predefined gene sets. Whether working at the group or individual level, the package offers a flexible and powerful tool for examining associations between brain imaging phenotypes (e.g., cortical thickness) and gene expression, using a variety of statistical models, null models, and aggregation methods.
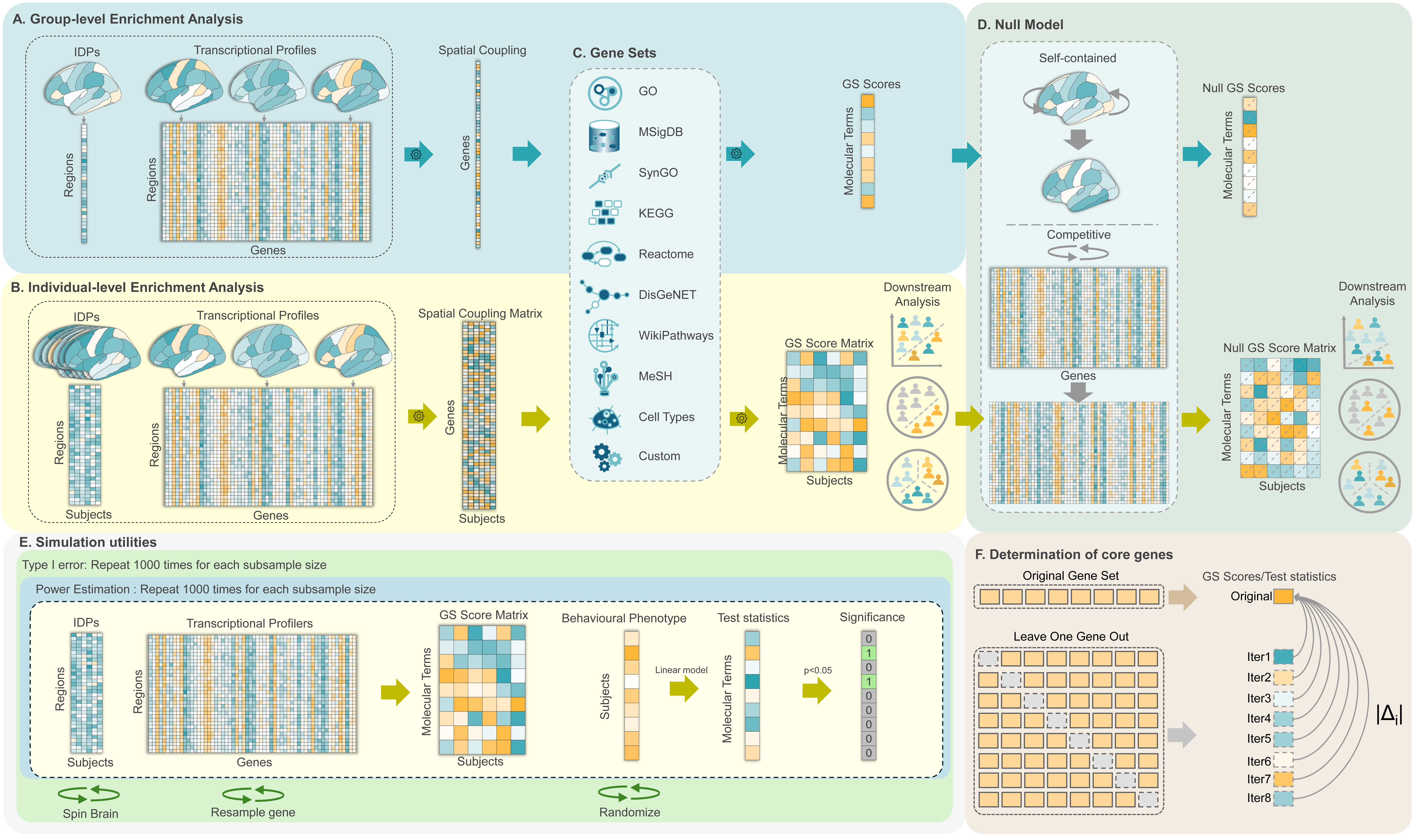
Overview of the BrainEnrich package and its analysis workflow.
🚀Features
- A. Group-Level Enrichment Analysis: A group-level IDP (e.g., effect size maps) is correlated with AHBA transcriptional profiles, yielding a spatial coupling vector that is aggregated within the predefined gene sets. The resultant “GS score” vector is tested against the null GS scores generated by the null models.
- B. Individual-Level Enrichment Analysis: A similar GS scoring approach is applied to individual IDPs, generating a spatial coupling matrix that is aggregated within the predefined gene sets. The resultant participant-specific GS scores are used for downstream analyses, and the results are tested against the null GS scores generated by the null models.
- C. Predefined Gene Sets: Predefined gene sets from various sources and user-defined sets aggregate gene-specific associations into GS scores for either group-level or individual-level analyses. The package includes predefined gene sets like: Gene Ontology, DisGeNet, KEGG, WikiPathways, Reactome, MeSH, SynGO, and Cell Type gene sets.
- D. Null Models: Includes both self-contained (spinning brain regions) and competitive (resampling genes) null models to assess the significance of GS score or to generate null GS scores at the individual level for downstream analysis.
- E. Simulation Studies: Type I error and power simulations to assess the reliability of the analysis methods.
- F. Core Genes Identification: A leave-one-out (LOO) procedure identifies genes that substantially influence GS scores (in group-level analysis) or the resultant test statistic (in individual-level analysis), highlighting the primary contributors to enriched terms.
- 🔧Multiple Association Methods: Pearson and Spearman correlations, Partial Least Squares (PLS) regression, and user-defined methods for exploring gene-imaging associations.
- 🔧Aggregation Methods: Multiple options for aggregating gene set scores, including mean, median, and Kolmogorov-Smirnov (KS)-based statistics.
💾Installation
# Install remotes if you haven't already
if (!requireNamespace("remotes", quietly = TRUE)) {
install.packages("remotes")}
if (!requireNamespace("BiocManager", quietly = TRUE)) {
install.packages("BiocManager")
}
BiocManager::install("DOSE")
# Install brainEnrich from GitHub
remotes::install_github("zh1peng/BrainEnrich")